Somatic Mutations in miRNA Genes in Lung Cancer-Potential Functional Consequences of Non-Coding Sequence Variants
- PMID: 31181801
- PMCID: PMC6627760
- DOI: 10.3390/cancers11060793
Somatic Mutations in miRNA Genes in Lung Cancer-Potential Functional Consequences of Non-Coding Sequence Variants
Abstract
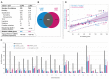
A growing body of evidence indicates that miRNAs may either drive or suppress oncogenesis. However, little is known about somatic mutations in miRNA genes. To determine the frequency and potential consequences of miRNA gene mutations, we analyzed whole exome sequencing datasets of 569 lung adenocarcinoma (LUAD) and 597 lung squamous cell carcinoma (LUSC) samples generated in The Cancer Genome Atlas (TCGA) project. Altogether, we identified 1091 somatic sequence variants affecting 522 different miRNA genes and showed that half of all cancers had at least one such somatic variant/mutation. These sequence variants occurred in most crucial parts of miRNA precursors, including mature miRNA and seed sequences. Due to our findings, we hypothesize that seed mutations may affect miRNA:target interactions, drastically changing the pool of predicted targets. Mutations may also affect miRNA biogenesis by changing the structure of miRNA precursors, DROSHA and DICER cleavage sites, and regulatory sequence/structure motifs. We identified 10 significantly overmutated hotspot miRNA genes, including the miR-379 gene in LUAD enriched in mutations in the mature miRNA and regulatory sequences. The occurrence of mutations in the hotspot miRNA genes was also shown experimentally. We present a comprehensive analysis of somatic variants in miRNA genes and show that some of these genes are mutational hotspots, suggesting their potential role in cancer.
Keywords: TCGA; lung cancer; miRNA; non-coding; somatic mutations.
Conflict of interest statement
The authors declare no conflicts of interest. The original manuscript has been uploaded to the bioRxiv preprint repository (doi: https://doi.org/10.1101/579011).
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

