Dynamic genome-scale cell-specific metabolic models reveal novel inter-cellular and intra-cellular metabolic communications during ovarian follicle development
- PMID: 31182013
- PMCID: PMC6558917
- DOI: 10.1186/s12859-019-2825-2
Dynamic genome-scale cell-specific metabolic models reveal novel inter-cellular and intra-cellular metabolic communications during ovarian follicle development
Abstract
Background: The maturation of the female germ cell, the oocyte, requires the synthesis and storing of all the necessary metabolites to support multiple divisions after fertilization. Oocyte maturation is only possible in the presence of surrounding, diverse, and changing layers of somatic cells. Our understanding of metabolic interactions between the oocyte and somatic cells has been limited due to dynamic nature of ovarian follicle development, thus warranting a systems approach.
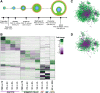
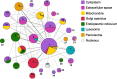
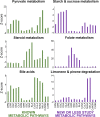
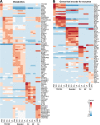
Results: Here, we developed a genome-scale metabolic model of the mouse ovarian follicle. This model was constructed using an updated mouse general metabolic model (Mouse Recon 2) and contains several key ovarian follicle development metabolic pathways. We used this model to characterize the changes in the metabolism of each follicular cell type (i.e., oocyte, granulosa cells, including cumulus and mural cells), during ovarian follicle development in vivo. Using this model, we predicted major metabolic pathways that are differentially active across multiple follicle stages. We identified a set of possible secreted and consumed metabolites that could potentially serve as biomarkers for monitoring follicle development, as well as metabolites for addition to in vitro culture media that support the growth and maturation of primordial follicles.
Conclusions: Our systems approach to model follicle metabolism can guide future experimental studies to validate the model results and improve oocyte maturation approaches and support growth of primordial follicles in vitro.
Keywords: Cell-type specific metabolic models; Genome-scale modeling; Metabolic communities; Metabolism; Ovarian follicle development; Secreted metabolites.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Ligands, Receptors, and Transcription Factors that Mediate Inter-Cellular and Intra-Cellular Communication during Ovarian Follicle Development.Reprod Sci. 2020 Feb;27(2):690-703. doi: 10.1007/s43032-019-00075-8. Epub 2020 Jan 14. Reprod Sci. 2020. PMID: 31939199 Free PMC article.
-
Control of Oocyte Growth and Development by Intercellular Communication Within the Follicular Niche.Results Probl Cell Differ. 2016;58:191-224. doi: 10.1007/978-3-319-31973-5_8. Results Probl Cell Differ. 2016. PMID: 27300180 Review.
-
Spatial Characterization of Bioenergetics and Metabolism of Primordial to Preovulatory Follicles in Whole Ex Vivo Murine Ovary.Biol Reprod. 2016 Dec;95(6):129. doi: 10.1095/biolreprod.116.142141. Epub 2016 Sep 28. Biol Reprod. 2016. PMID: 27683265 Free PMC article.
-
Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors.Hum Reprod Update. 2016 Dec;23(1):1-18. doi: 10.1093/humupd/dmw039. Epub 2016 Oct 26. Hum Reprod Update. 2016. PMID: 27797914 Free PMC article. Review.
-
Differences in cumulus cell gene expression indicate the benefit of a pre-maturation step to improve in-vitro bovine embryo production.Mol Hum Reprod. 2016 Dec;22(12):882-897. doi: 10.1093/molehr/gaw055. Epub 2016 Aug 24. Mol Hum Reprod. 2016. PMID: 27559149
Cited by
-
Ligands, Receptors, and Transcription Factors that Mediate Inter-Cellular and Intra-Cellular Communication during Ovarian Follicle Development.Reprod Sci. 2020 Feb;27(2):690-703. doi: 10.1007/s43032-019-00075-8. Epub 2020 Jan 14. Reprod Sci. 2020. PMID: 31939199 Free PMC article.
-
Metabolic Control of Germline Formation and Differentiation in Mammals.Sex Dev. 2022;16(5-6):388-403. doi: 10.1159/000520662. Epub 2022 Jan 27. Sex Dev. 2022. PMID: 35086109 Free PMC article. Review.
-
Successful 3D culture and transplantation of mouse isolated preantral follicles in hydrogel of bioengineered Wharton's jelly.PLoS One. 2023 Sep 20;18(9):e0290095. doi: 10.1371/journal.pone.0290095. eCollection 2023. PLoS One. 2023. PMID: 37729236 Free PMC article.
-
Correlation between Pre-Ovulatory Follicle Diameter and Follicular Fluid Metabolome Profiles in Lactating Beef Cows.Metabolites. 2021 Sep 14;11(9):623. doi: 10.3390/metabo11090623. Metabolites. 2021. PMID: 34564438 Free PMC article.
-
Multi-omics reveal the metabolic patterns in mouse cumulus cells during oocyte maturation.J Ovarian Res. 2023 Aug 8;16(1):156. doi: 10.1186/s13048-023-01237-8. J Ovarian Res. 2023. PMID: 37550748 Free PMC article.
References
-
- Sugiura K, Pendola FL, Eppig JJ. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev Biol. 2005;279(1):20–30. - PubMed
-
- Eppig JJ, Pendola FL, Wigglesworth K, Pendola JK. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biol Reprod. 2005;73(2):351–357. - PubMed
-
- Downs SM, Verhoeven AD. Glutamine and the maintenance of meiotic arrest in mouse oocytes: influence of culture medium, glucose, and cumulus cells. Biol Reprod. 2002;66:116. - PubMed
-
- Downs SM, Daniel SAJ, Bornslaeger EA, Hoppe PC, Eppig JJ. Maintenance of meiotic arrest in mouse oocytes by purines - modulation of camp levels and camp phosphodiesterase activity. Gamete Res. 1989;23(3):323–334. - PubMed

