Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale
- PMID: 31182022
- PMCID: PMC6558717
- DOI: 10.1186/s12870-019-1851-6
Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale
Abstract
Background: Studies have demonstrated that BBX (B-BOX) genes play crucial roles in regulatory networks controlling plant growth, developmental processes and stress response. Nevertheless, comprehensive study of BBX genes in orchids (Orchidaceae) is not well studied. The newly released genome sequences of Dendrobium officinale and Phalaenopsis equestris have allowed a systematic analysis of these important BBX genes in orchids.
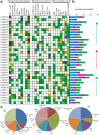
Results: Here we identified 19 (DoBBX01-19) and 16 (PeBBX01-16) BBX genes from D. officinale and P. equestris, respectively, and clustered into five clades (I-V) according to phylogenetic analysis. Thirteen orthologous, two DoBBXs paralogous and two PeBBXs paralogous gene pairs were validated. This gene family mainly underwent purifying selection, but five domains experienced positive selection during evolution. Noteworthy, the expression patterns of root, root_tips, stem, leaf, speal, column, lip, and flower_buds revealed that they might contribution to the formation of these tissues. According to the cis-regulatory elements analysis of BBX genes, qRT-PCR experiments were carried out using D. officinale PLBs (protocorm-like bodies) and displayed that these BBX genes were differentially regulated under AgNO3, MeJA (Methyl Jasmonate), ABA (abscisic acid) and SA (salicylic acid) treatments.
Conclusions: Our analysis exposed that DoBBX genes play significant roles in plant growth and development, and response to different environmental stress conditions of D. officinale, which provide aid in the selection of appropriate candidate genes for further functional characterization of BBX genes in plants.
Keywords: Abiotic stress; BBX; D. officinale; Evolution; qRT-PCR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Molecular characterization and expression analysis of WRKY family genes in Dendrobium officinale.Genes Genomics. 2018 Mar;40(3):265-279. doi: 10.1007/s13258-017-0602-z. Epub 2017 Nov 17. Genes Genomics. 2018. PMID: 29892797
-
Genome-wide identification of Ankyrin (ANK) repeat gene families in three Dendrobium species and the expression of ANK genes in D. officinale under gibberellin and abscisic acid treatments.BMC Plant Biol. 2024 Aug 10;24(1):762. doi: 10.1186/s12870-024-05461-2. BMC Plant Biol. 2024. PMID: 39123107 Free PMC article.
-
Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale.Int J Mol Sci. 2021 May 14;22(10):5221. doi: 10.3390/ijms22105221. Int J Mol Sci. 2021. PMID: 34069261 Free PMC article.
-
[BBX transcriptional factors family in plants-a review].Sheng Wu Gong Cheng Xue Bao. 2020 Apr 25;36(4):666-677. doi: 10.13345/j.cjb.190302. Sheng Wu Gong Cheng Xue Bao. 2020. PMID: 32347061 Review. Chinese.
-
The B-box bridge between light and hormones in plants.J Photochem Photobiol B. 2019 Feb;191:164-174. doi: 10.1016/j.jphotobiol.2018.12.021. Epub 2018 Dec 28. J Photochem Photobiol B. 2019. PMID: 30640143 Review.
Cited by
-
Genome-Wide Characterization and Expression Analysis of HD-ZIP Gene Family in Dendrobium officinale.Front Genet. 2022 Mar 18;13:797014. doi: 10.3389/fgene.2022.797014. eCollection 2022. Front Genet. 2022. PMID: 35368655 Free PMC article.
-
Network Analysis of Transcriptome and LC-MS Reveals a Possible Biosynthesis Pathway of Anthocyanins in Dendrobium officinale.Biomed Res Int. 2020 Apr 29;2020:6512895. doi: 10.1155/2020/6512895. eCollection 2020. Biomed Res Int. 2020. PMID: 32420359 Free PMC article.
-
Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana.Plant Signal Behav. 2020 Sep 1;15(9):1782647. doi: 10.1080/15592324.2020.1782647. Epub 2020 Jun 17. Plant Signal Behav. 2020. PMID: 32552524 Free PMC article.
-
Divergence of flowering-related genes to control flowering in five Euphorbiaceae genomes.Front Plant Sci. 2022 Oct 19;13:1015114. doi: 10.3389/fpls.2022.1015114. eCollection 2022. Front Plant Sci. 2022. PMID: 36340397 Free PMC article.
-
Comparative Analysis and Expression Patterns of the PLP_deC Genes in Dendrobium officinale.Int J Mol Sci. 2019 Dec 20;21(1):54. doi: 10.3390/ijms21010054. Int J Mol Sci. 2019. PMID: 31861760 Free PMC article.
References
-
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80(6):847. - PubMed
-
- Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. Faseb J. 1995;9(8):597–604. - PubMed
-
- Torok M, Etkin LD. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. 2001. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

