Early Notch Signals Induce a Pathogenic Molecular Signature during Priming of Alloantigen-Specific Conventional CD4+ T Cells in Graft-versus-Host Disease
- PMID: 31182480
- PMCID: PMC6615974
- DOI: 10.4049/jimmunol.1900192
Early Notch Signals Induce a Pathogenic Molecular Signature during Priming of Alloantigen-Specific Conventional CD4+ T Cells in Graft-versus-Host Disease
Abstract
Graft-versus-host disease (GVHD) is the most serious complication of allogeneic hematopoietic cell transplantation. Notch signals delivered during the first 48 h after transplantation drive proinflammatory cytokine production in conventional T cells (Tconv) and inhibit the expansion of regulatory T cells (Tregs). Short-term Notch inhibition induces long-term GVHD protection. However, it remains unknown whether Notch blockade blunts GVHD through its effects on Tconv, Tregs, or both and what early Notch-regulated molecular events occur in alloantigen-specific T cells. To address these questions, we engineered T cell grafts to achieve selective Notch blockade in Tconv versus Tregs and evaluated their capacity to trigger GVHD in mice. Notch blockade in Tconv was essential for GVHD protection as GVHD severity was similar in the recipients of wild-type Tconv combined with Notch-deprived versus wild-type Tregs. To identify the impact of Notch signaling on the earliest steps of T cell activation in vivo, we established a new acute GVHD model mediated by clonal alloantigen-specific 4C CD4+ Tconv. Notch-deprived 4C T cells had preserved early steps of activation, IL-2 production, proliferation, and Th cell polarization. In contrast, Notch inhibition dampened IFN-γ and IL-17 production, diminished mTORC1 and ERK1/2 activation, and impaired transcription of a subset of Myc-regulated genes. The distinct Notch-regulated signature had minimal overlap with known Notch targets in T cell leukemia and developing T cells, highlighting the specific impact of Notch signaling in mature T cells. Our findings uncover a unique molecular program associated with the pathogenic effects of Notch in T cells at the earliest stages of GVHD.
Copyright © 2019 by The American Association of Immunologists, Inc.
Figures
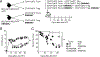

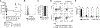
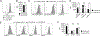
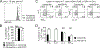


References
-
- Radtke F, Fasnacht N, and Macdonald HR. 2010. Notch signaling in the immune system. Immunity 32: 14–27. - PubMed
-
- Zhang Y, Sandy AR, Wang J, Radojcic V, Shan GT, Tran IT, Friedman A, Kato K, He S, Cui S, Hexner E, Frank DM, Emerson SG, Pear WS, and Maillard I. 2011. Notch signaling is a critical regulator of allogeneic CD4+ T-cell responses mediating graft-versus-host disease. Blood 117: 299–308. - PMC - PubMed
-
- Mochizuki K, Xie F, He S, Tong Q, Liu Y, Mochizuki I, Guo Y, Kato K, Yagita H, Mineishi S, and Zhang Y. 2013. Delta-like ligand 4 identifies a previously uncharacterized population of inflammatory dendritic cells that plays important roles in eliciting allogeneic T cell responses in mice. J Immunol 190: 3772–3782. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

