PhoP-Mediated Repression of the SPI1 Type 3 Secretion System in Salmonella enterica Serovar Typhimurium
- PMID: 31182495
- PMCID: PMC6657598
- DOI: 10.1128/JB.00264-19
PhoP-Mediated Repression of the SPI1 Type 3 Secretion System in Salmonella enterica Serovar Typhimurium
Abstract
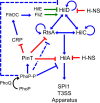
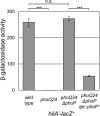
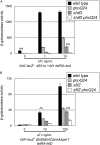
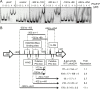
Salmonella must rapidly adapt to various niches in the host during infection. Relevant virulence factors must be appropriately induced, and systems that are detrimental in a particular environment must be turned off. Salmonella infects intestinal epithelial cells using a type 3 secretion system (T3SS) encoded on Salmonella pathogenicity island 1 (SPI1). The system is controlled by three AraC-like regulators, HilD, HilC, and RtsA, which form a complex feed-forward loop to activate expression of hilA, encoding the main transcriptional regulator of T3SS structural genes. This system is tightly regulated, with many of the activating signals acting at the level of hilD translation or HilD protein activity. Once inside the phagosomes of epithelial cells, or in macrophages during systemic stages of disease, the SPI1 T3SS is no longer required or expressed. Here, we show that the PhoPQ two-component system, critical for intracellular survival, appears to be the primary mechanism by which Salmonella shuts down the SPI1 T3SS. PhoP negatively regulates hilA through multiple distinct mechanisms: direct transcriptional repression of the hilA promoter, indirect transcriptional repression of both the hilD and rtsA promoters, and activation of the small RNA (sRNA) PinT. Genetic analyses and electrophoretic mobility shift assays suggest that PhoP specifically binds the hilA promoter to block binding of activators HilD, HilC, and RtsA as a mechanism of repression.IMPORTANCESalmonella is one of the most common foodborne pathogens, causing an estimated 1.2 million illnesses per year in the United States. A key step in infection is the activation of the bacterial invasion machinery, which induces uptake of the bacterium into epithelial cells and leads to induction of inflammatory diarrhea. Upon entering the vacuolar compartments of host cells, Salmonella senses an environmental transition and represses the invasion machinery with a two-component system relevant for survival within the vacuole. This adaptation to specific host niches is an important example of how signals are integrated for survival of the pathogen.
Keywords: PhoPQ; Salmonella; virulence regulation.
Copyright © 2019 American Society for Microbiology.
Figures


Similar articles
-
The Small RNA PinT Contributes to PhoP-Mediated Regulation of the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2019 Sep 6;201(19):e00312-19. doi: 10.1128/JB.00312-19. Print 2019 Oct 1. J Bacteriol. 2019. PMID: 31262841 Free PMC article.
-
HilD, HilC, and RtsA Form Homodimers and Heterodimers To Regulate Expression of the Salmonella Pathogenicity Island I Type III Secretion System.J Bacteriol. 2020 Apr 9;202(9):e00012-20. doi: 10.1128/JB.00012-20. Print 2020 Apr 9. J Bacteriol. 2020. PMID: 32041797 Free PMC article.
-
The Small RNA MicC Downregulates hilD Translation To Control the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2022 Jan 18;204(1):e0037821. doi: 10.1128/JB.00378-21. Epub 2021 Oct 25. J Bacteriol. 2022. PMID: 34694902 Free PMC article.
-
Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium.Curr Opin Microbiol. 2007 Feb;10(1):24-9. doi: 10.1016/j.mib.2006.12.002. Epub 2007 Jan 5. Curr Opin Microbiol. 2007. PMID: 17208038 Review.
-
Salmonella invasion gene regulation: a story of environmental awareness.J Microbiol. 2005 Feb;43 Spec No:110-7. J Microbiol. 2005. PMID: 15765064 Review.
Cited by
-
Functions of Small Non-Coding RNAs in Salmonella-Host Interactions.Biology (Basel). 2022 Aug 29;11(9):1283. doi: 10.3390/biology11091283. Biology (Basel). 2022. PMID: 36138763 Free PMC article. Review.
-
An Advanced Human Intestinal Coculture Model Reveals Compartmentalized Host and Pathogen Strategies during Salmonella Infection.mBio. 2020 Feb 18;11(1):e03348-19. doi: 10.1128/mBio.03348-19. mBio. 2020. PMID: 32071273 Free PMC article.
-
Type III Secretion Effectors with Arginine N-Glycosyltransferase Activity.Microorganisms. 2020 Mar 2;8(3):357. doi: 10.3390/microorganisms8030357. Microorganisms. 2020. PMID: 32131463 Free PMC article. Review.
-
Long-Distance Effects of H-NS Binding in the Control of hilD Expression in the Salmonella SPI1 Locus.J Bacteriol. 2021 Oct 12;203(21):e0030821. doi: 10.1128/JB.00308-21. Epub 2021 Aug 23. J Bacteriol. 2021. PMID: 34424033 Free PMC article.
-
An incoherent feedforward loop formed by SirA/BarA, HilE and HilD is involved in controlling the growth cost of virulence factor expression by Salmonella Typhimurium.PLoS Pathog. 2021 May 28;17(5):e1009630. doi: 10.1371/journal.ppat.1009630. eCollection 2021 May. PLoS Pathog. 2021. PMID: 34048498 Free PMC article.
References
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi:10.1086/650733. - DOI - PubMed
-
- Zhou D, Chen LM, Hernandez L, Shears SB, Galan JE. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol Microbiol 39:248–259. doi:10.1046/j.1365-2958.2001.02230.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

