Sequence and structural patterns detected in entangled proteins reveal the importance of co-translational folding
- PMID: 31182755
- PMCID: PMC6557820
- DOI: 10.1038/s41598-019-44928-3
Sequence and structural patterns detected in entangled proteins reveal the importance of co-translational folding
Abstract
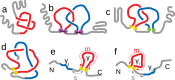
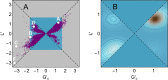
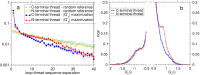
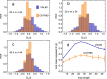
Proteins must fold quickly to acquire their biologically functional three-dimensional native structures. Hence, these are mainly stabilized by local contacts, while intricate topologies such as knots are rare. Here, we reveal the existence of specific patterns adopted by protein sequences and structures to deal with backbone self-entanglement. A large scale analysis of the Protein Data Bank shows that loops significantly intertwined with another chain portion are typically closed by weakly bound amino acids. Why is this energetic frustration maintained? A possible picture is that entangled loops are formed only toward the end of the folding process to avoid kinetic traps. Consistently, these loops are more frequently found to be wrapped around a portion of the chain on their N-terminal side, the one translated earlier at the ribosome. Finally, these motifs are less abundant in natural native states than in simulated protein-like structures, yet they appear in 32% of proteins, which in some cases display an amazingly complex intertwining.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Dill K, MacCallum J. The protein-folding problem. 50 years on. Science. 2012;338:1042–1046. - PubMed
-
- Micheletti C, Banavar JR, Maritan A, Seno F. Protein structures and optimal folding from a geometrical variational principle. Physical Review Letters. 1999;82:3372. doi: 10.1103/PhysRevLett.82.3372. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

