DNA-uptake pili of Vibrio cholerae are required for chitin colonization and capable of kin recognition via sequence-specific self-interaction
- PMID: 31182799
- PMCID: PMC6708440
- DOI: 10.1038/s41564-019-0479-5
DNA-uptake pili of Vibrio cholerae are required for chitin colonization and capable of kin recognition via sequence-specific self-interaction
Abstract
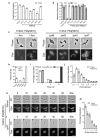
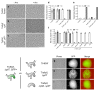
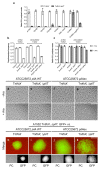
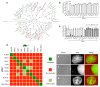
How bacteria colonize surfaces and how they distinguish the individuals around them are fundamental biological questions. Type IV pili are a widespread and multipurpose class of cell surface polymers. Here we directly visualize the DNA-uptake pilus of Vibrio cholerae, which is produced specifically during growth on its natural habitat-chitinous surfaces. As predicted, these pili are highly dynamic and retract before DNA uptake during competence for natural transformation. Interestingly, DNA-uptake pili can also self-interact to mediate auto-aggregation. This capability is conserved in disease-causing pandemic strains, which typically encode the same major pilin subunit, PilA. Unexpectedly, however, we discovered that extensive strain-to-strain variability in PilA (present in environmental isolates) creates a set of highly specific interactions, enabling cells producing pili composed of different PilA subunits to distinguish between one another. We go on to show that DNA-uptake pili bind to chitinous surfaces and are required for chitin colonization under flow, and that pili capable of self-interaction connect cells on chitin within dense pili networks. Our results suggest a model whereby DNA-uptake pili function to promote inter-bacterial interactions during surface colonization. Moreover, they provide evidence that type IV pili could offer a simple and potentially widespread mechanism for bacterial kin recognition.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
Settling down on chitin.Nat Rev Microbiol. 2019 Sep;17(9):527. doi: 10.1038/s41579-019-0237-y. Nat Rev Microbiol. 2019. PMID: 31289382 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

