Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders
- PMID: 31182824
- PMCID: PMC6831729
- DOI: 10.1038/s41436-019-0554-6
Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders
Erratum in
-
Correction: Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders.Genet Med. 2020 Oct;22(10):1731-1732. doi: 10.1038/s41436-020-0913-3. Genet Med. 2020. PMID: 32728138 Free PMC article.
Abstract
Purpose: For neurodevelopmental disorders (NDDs), etiological evaluation can be a diagnostic odyssey involving numerous genetic tests, underscoring the need to develop a streamlined algorithm maximizing molecular diagnostic yield for this clinical indication. Our objective was to compare the yield of exome sequencing (ES) with that of chromosomal microarray (CMA), the current first-tier test for NDDs.
Methods: We performed a PubMed scoping review and meta-analysis investigating the diagnostic yield of ES for NDDs as the basis of a consensus development conference. We defined NDD as global developmental delay, intellectual disability, and/or autism spectrum disorder. The consensus development conference included input from genetics professionals, pediatric neurologists, and developmental behavioral pediatricians.
Results: After applying strict inclusion/exclusion criteria, we identified 30 articles with data on molecular diagnostic yield in individuals with isolated NDD, or NDD plus associated conditions (such as Rett-like features). Yield of ES was 36% overall, 31% for isolated NDD, and 53% for the NDD plus associated conditions. ES yield for NDDs is markedly greater than previous studies of CMA (15-20%).
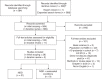
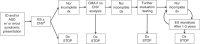
Conclusion: Our review demonstrates that ES consistently outperforms CMA for evaluation of unexplained NDDs. We propose a diagnostic algorithm placing ES at the beginning of the evaluation of unexplained NDDs.
Keywords: autism; consensus development conference; diagnostic yield; genetic testing; intellectual disability.
Conflict of interest statement
All authors completed the International Committee of Medical Journal Editors conflict of interest disclosure forms, which were reviewed by the senior authors. H.V.F. reports personal fees from UpToDate, outside the submitted work. T.F. has received federal funding or research support from, acted as a consultant to, received travel support from, and/or received a speaker's honorarium from the Cole Family Research Fund, Simons Foundation, Ingalls Foundation, Forest Laboratories, Ecoeos, IntegraGen, Kugona LLC, Shire Development, Bristol-Myers Squibb, Roche Pharma, National Institutes of Health, and the Brain and Behavior Research Foundation, all unrelated to this project. S.W.S. is on the Scientific Advisory Committees of Population Bio and Deep Genomics, and intellectual property from his research held at the Hospital for Sick Children is licensed to Athena Diagnostics, and separately Lineagen. These relationships did not influence data interpretation or presentation during this study, but are being disclosed for potential future considerations. M.S. reports grant support from Novartis, Roche, Pfizer, Ipsen, LAM Therapeutics and Quadrant Biosciences, and he has served on Scientific Advisory Boards for Sage, Roche, Celgene, and Takeda, all unrelated to this project. The other authors declare no conflicts of interest.
Figures

References
-
- Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. - PubMed
-
- Schaefer GB, Mendelsohn NJ, Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407. - PubMed
-
- Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18. - PubMed

