Zika virus antagonizes interferon response in patients and disrupts RIG-I-MAVS interaction through its CARD-TM domains
- PMID: 31183075
- PMCID: PMC6555941
- DOI: 10.1186/s13578-019-0308-9
Zika virus antagonizes interferon response in patients and disrupts RIG-I-MAVS interaction through its CARD-TM domains
Abstract
Background: The emerging threat to global health associated with the Zika virus (ZIKV) epidemics and its link to severe complications highlights a growing need to better understand the pathogenic mechanisms of ZIKV. Accumulating evidence for a critical role of type I interferon (IFN-I) in protecting hosts from ZIKV infection lies in the findings that ZIKV has evolved various strategies to subvert the host defense line by counteracting the early IFN induction or subsequent IFN signaling. Yet, mechanisms underlying the counter-IFN capability of ZIKV and its proteins, which might contribute to the well-recognized broad cellular tropisms and persistence of ZIKV, remain incompletely understood.
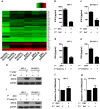
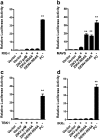
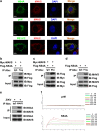
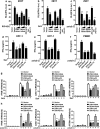
Results: Using RNA sequencing-based transcriptional profiling of whole blood cells isolated from patients acutely infected by ZIKV, we found that transcriptional signature programs of antiviral interferon-stimulated genes and innate immune sensors in ZIKV-infected patients remained inactive as compared to those of healthy donors, suggesting that ZIKV was able to suppress the induction of IFN-I during the natural infection process in humans. Furthermore, by analyzing the molecular interaction in a ZIKV NS4A-overexpression system, or in the context of actual ZIKV infection, we identified that ZIKV NS4A directly bound MAVS and thereby interrupted the RIG-I/MAVS interaction through the CARD-TM domains, leading to attenuated production of IFN-I.
Conclusions: Our findings collectively revealed that ZIKV NS4A targeted MAVS and contributed to ZIKV immune evasion through abrogating MAVS-mediated IFN production. These findings obtained from patient studies have added new knowledge and molecular details to our understanding regarding how ZIKV mediates suppression of the IFN-I system and may provide a new basis for the future development of anti-ZIKV strategies.
Keywords: Interferon; MAVS; Nonstructural protein 4A; RIG-I; Zika virus.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures

Similar articles
-
Zika Virus Non-structural Protein 4A Blocks the RLR-MAVS Signaling.Front Microbiol. 2018 Jun 25;9:1350. doi: 10.3389/fmicb.2018.01350. eCollection 2018. Front Microbiol. 2018. PMID: 29988497 Free PMC article.
-
Dengue Virus Subverts Host Innate Immunity by Targeting Adaptor Protein MAVS.J Virol. 2016 Jul 27;90(16):7219-7230. doi: 10.1128/JVI.00221-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27252539 Free PMC article.
-
Zika Virus Proteins NS2A and NS4A Are Major Antagonists that Reduce IFN-β Promoter Activity Induced by the MDA5/RIG-I Signaling Pathway.J Microbiol Biotechnol. 2019 Oct 28;29(10):1665-1674. doi: 10.4014/jmb.1909.09017. J Microbiol Biotechnol. 2019. PMID: 31581385
-
Immune Recognition versus Immune Evasion Systems in Zika Virus Infection.Biomedicines. 2023 Feb 20;11(2):642. doi: 10.3390/biomedicines11020642. Biomedicines. 2023. PMID: 36831177 Free PMC article. Review.
-
Running interference: Interplay between Zika virus and the host interferon response.Cytokine. 2019 Jul;119:7-15. doi: 10.1016/j.cyto.2019.02.009. Epub 2019 Mar 8. Cytokine. 2019. PMID: 30856603 Review.
Cited by
-
Zika virus impacts extracellular vesicle composition and cellular gene expression in macaque early gestation trophoblasts.Sci Rep. 2022 May 5;12(1):7348. doi: 10.1038/s41598-022-11275-9. Sci Rep. 2022. PMID: 35513694 Free PMC article.
-
Interaction between NS1 and Cellular MAVS Contributes to NS1 Mitochondria Targeting.Viruses. 2021 Sep 23;13(10):1909. doi: 10.3390/v13101909. Viruses. 2021. PMID: 34696339 Free PMC article.
-
Single Amino Acid Mutations Affect Zika Virus Replication In Vitro and Virulence In Vivo.Viruses. 2020 Nov 12;12(11):1295. doi: 10.3390/v12111295. Viruses. 2020. PMID: 33198111 Free PMC article.
-
Innate Immune Response to Powassan Virus Infection: Progress Toward Infection Control.Vaccines (Basel). 2025 Jul 15;13(7):754. doi: 10.3390/vaccines13070754. Vaccines (Basel). 2025. PMID: 40733731 Free PMC article. Review.
-
Mosquito-borne flaviviruses and type I interferon: catch me if you can!Front Microbiol. 2023 Oct 30;14:1257024. doi: 10.3389/fmicb.2023.1257024. eCollection 2023. Front Microbiol. 2023. PMID: 37965539 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

