The innate immune sensor Toll-like receptor 2 controls the senescence-associated secretory phenotype
- PMID: 31183403
- PMCID: PMC6551188
- DOI: 10.1126/sciadv.aaw0254
The innate immune sensor Toll-like receptor 2 controls the senescence-associated secretory phenotype
Abstract
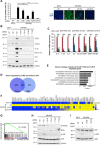
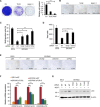
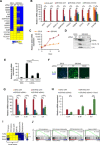
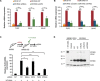
Cellular senescence is a stress response program characterized by a robust cell cycle arrest and the induction of a proinflammatory senescence-associated secretory phenotype (SASP) that is triggered through an unknown mechanism. Here, we show that, during oncogene-induced senescence (OIS), the Toll-like receptor 2 (TLR2) and its partner TLR10 are key mediators of senescence in vitro and in murine models. TLR2 promotes cell cycle arrest by regulating the tumor suppressors p53-p21CIP1, p16INK4a, and p15INK4b and regulates the SASP through the induction of the acute-phase serum amyloids A1 and A2 (A-SAAs) that, in turn, function as the damage-associated molecular patterns (DAMPs) signaling through TLR2 in OIS. Last, we found evidence that the cGAS-STING cytosolic DNA sensing pathway primes TLR2 and A-SAAs expression in OIS. In summary, we report that innate immune sensing of senescence-associated DAMPs by TLR2 controls the SASP and reinforces the cell cycle arrest program in OIS.
Figures


References
-
- Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W., Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997). - PubMed
-
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A. H. F. M., Schlegelberger B., Stein H., Dörken B., Jenuwein T., Schmitt C. A., Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005). - PubMed
-
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., Beach D., Serrano M., Tumour biology: Senescence in premalignant tumours. Nature 436, 642 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

