Connections between metabolism and epigenetics in cancers
- PMID: 31185282
- PMCID: PMC6690786
- DOI: 10.1016/j.semcancer.2019.06.006
Connections between metabolism and epigenetics in cancers
Abstract
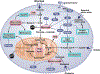
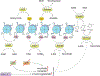
In the past half century, our version on cancer, from tumor initiation, growth, to metastasis, is dominated by genetic mutation. The importance of metabolism and epigenetics was not recognized until most recently. Extensive cell proliferation is one of the hallmarks of cancers. To support the energetic and anabolic demands of enhanced proliferation, tumors reprogram the pathways of nutrient procurement and metabolism. In this context, a new link between metabolic alterations and cancer progression has been unraveled over the last decade by the studies conducted in the area of cancer cell metabolism. Cancer cells are known to alter their metabolic profile during the course of tumorigenesis and metastasis thereby exhibiting a tightly regulated program of metabolic plasticity. Noteworthy, certain metabolic alteration are known to occur at the epigenetic level, thus making epigenetics and metabolism highly interwoven in a reciprocal manner. Metabolites that are generated during metabolic pathways, such as in glycolytic cycle and oxidative phosphorylation, serve as cofactors or substrates for the enzymatic reactions that catalyze the epigenetic modifications and transcriptional regulation. Several studies also indicate that the epigenome is sensitive to cellular metabolism. Since many of the metabolic alterations and consequently aberrated epigenetic regulation are common to a wide range of cancer types, they serve as promising targets for anti-cancer therapies. Here we discuss the latest findings in cancer cell metabolism, elucidating the major anabolic, catabolic and energetic demands required for sustaining cancer growth, and the influence of altered metabolism on epigenetics and vice versa. A comprehensive research pertaining to metabolomic profiling and epigenome interactors/mediators in malignant neoplasias is imperative in deciphering the potential targets that can be exploited for the development of robust anti-cancer therapies.
Keywords: Cancer cell metabolism; DNA and histone methylation; Epigenetics; Warburg effect.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures
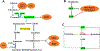
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

