Metaphenomic Responses of a Native Prairie Soil Microbiome to Moisture Perturbations
- PMID: 31186334
- PMCID: PMC6561317
- DOI: 10.1128/mSystems.00061-19
Metaphenomic Responses of a Native Prairie Soil Microbiome to Moisture Perturbations
Abstract
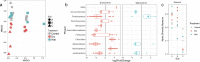
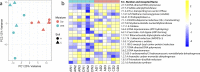
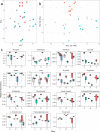
Climate change is causing shifts in precipitation patterns in the central grasslands of the United States, with largely unknown consequences on the collective physiological responses of the soil microbial community, i.e., the metaphenome. Here, we used an untargeted omics approach to determine the soil microbial community's metaphenomic response to soil moisture and to define specific metabolic signatures of the response. Specifically, we aimed to develop the technical approaches and metabolic mapping framework necessary for future systematic ecological studies. We collected soil from three locations at the Konza Long-Term Ecological Research (LTER) field station in Kansas, and the soils were incubated for 15 days under dry or wet conditions and compared to field-moist controls. The microbiome response to wetting or drying was determined by 16S rRNA amplicon sequencing, metatranscriptomics, and metabolomics, and the resulting shifts in taxa, gene expression, and metabolites were assessed. Soil drying resulted in significant shifts in both the composition and function of the soil microbiome. In contrast, there were few changes following wetting. The combined metabolic and metatranscriptomic data were used to generate reaction networks to determine the metaphenomic response to soil moisture transitions. Site location was a strong determinant of the response of the soil microbiome to moisture perturbations. However, some specific metabolic pathways changed consistently across sites, including an increase in pathways and metabolites for production of sugars and other osmolytes as a response to drying. Using this approach, we demonstrate that despite the high complexity of the soil habitat, it is possible to generate insight into the effect of environmental change on the soil microbiome and its physiology and functions, thus laying the groundwork for future, targeted studies.IMPORTANCE Climate change is predicted to result in increased drought extent and intensity in the highly productive, former tallgrass prairie region of the continental United States. These soils store large reserves of carbon. The decrease in soil moisture due to drought has largely unknown consequences on soil carbon cycling and other key biogeochemical cycles carried out by soil microbiomes. In this study, we found that soil drying had a significant impact on the structure and function of soil microbial communities, including shifts in expression of specific metabolic pathways, such as those leading toward production of osmoprotectant compounds. This study demonstrates the application of an untargeted multi-omics approach to decipher details of the soil microbial community's metaphenotypic response to environmental perturbations and should be applicable to studies of other complex microbial systems as well.
Keywords: metaphenome; metatranscriptome; multi-omics; soil microbiome.
Copyright © 2019 Roy Chowdhury et al.
Figures


References
-
- Cook BI, Seager R, Miller RL. 2011. Atmospheric circulation anomalies during two persistent North American droughts: 1932–1939 and 1948–1957. Clim Dyn 36:2339–2355. doi:10.1007/s00382-010-0807-1. - DOI
-
- Intergovernmental Panel on Climate Change. 2013. The physical science basis: working group I contribution to the fifth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, New York, NY.
-
- Porporato A, Vico G, Fay P. 2006. Superstatistics of hydro-climatic fluctuations and interannual ecosystem productivity. Geophys Res Lett 33:L15402. doi:10.1029/2006GL026412. - DOI
-
- Guo Y, Gong P, Amundson R, Yu Q. 2006. Analysis of factors controlling soil carbon in the conterminous United States. Soil Sci Soc Am J 70:601–612. doi:10.2136/sssaj2005.0163. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

