Multi-omics Signature of Candida auris, an Emerging and Multidrug-Resistant Pathogen
- PMID: 31186339
- PMCID: PMC6561322
- DOI: 10.1128/mSystems.00257-19
Multi-omics Signature of Candida auris, an Emerging and Multidrug-Resistant Pathogen
Abstract
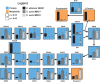
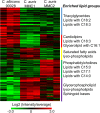
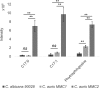
Candida auris is a recently described pathogenic fungus that is causing invasive outbreaks on all continents. The fungus is of high concern given the numbers of multidrug-resistant strains that have been isolated in distinct sites across the globe. The fact that its diagnosis is still problematic suggests that the spreading of the pathogen remains underestimated. Notably, the molecular mechanisms of virulence and antifungal resistance employed by this new species are largely unknown. In the present work, we compared two clinical isolates of C. auris with distinct drug susceptibility profiles and a Candida albicans reference strain using a multi-omics approach. Our results show that, despite the distinct drug resistance profile, both C. auris isolates appear to be very similar, albeit with a few notable differences. However, compared to C. albicans both C. auris isolates have major differences regarding their carbon utilization and downstream lipid and protein content, suggesting a multifactorial mechanism of drug resistance. The molecular profile displayed by C. auris helps to explain the antifungal resistance and virulence phenotypes of this new emerging pathogen.IMPORTANCE Candida auris was first described in Japan in 2009 and has now been the cause of significant outbreaks across the globe. The high number of isolates that are resistant to one or more antifungals, as well as the high mortality rates from patients with bloodstream infections, has attracted the attention of the medical mycology, infectious disease, and public health communities to this pathogenic fungus. In the current work, we performed a broad multi-omics approach on two clinical isolates isolated in New York, the most affected area in the United States and found that the omic profile of C. auris differs significantly from C. albicans In addition to our insights into C. auris carbon utilization and lipid and protein content, we believe that the availability of these data will enhance our ability to combat this rapidly emerging pathogenic yeast.
Keywords: Candida auris; antifungal resistance; fluconazole; multi-omics.
Copyright © 2019 Zamith-Miranda et al.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

