Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns
- PMID: 31186427
- PMCID: PMC6559955
- DOI: 10.1038/s41467-019-10461-0
Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns
Abstract
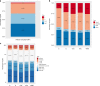
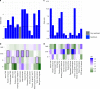
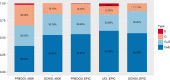
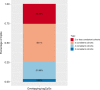
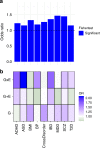
Epigenetic processes, including DNA methylation (DNAm), are among the mechanisms allowing integration of genetic and environmental factors to shape cellular function. While many studies have investigated either environmental or genetic contributions to DNAm, few have assessed their integrated effects. Here we examine the relative contributions of prenatal environmental factors and genotype on DNA methylation in neonatal blood at variably methylated regions (VMRs) in 4 independent cohorts (overall n = 2365). We use Akaike's information criterion to test which factors best explain variability of methylation in the cohort-specific VMRs: several prenatal environmental factors (E), genotypes in cis (G), or their additive (G + E) or interaction (GxE) effects. Genetic and environmental factors in combination best explain DNAm at the majority of VMRs. The CpGs best explained by either G, G + E or GxE are functionally distinct. The enrichment of genetic variants from GxE models in GWAS for complex disorders supports their importance for disease risk.
Conflict of interest statement
E.B.B. is co-inventor on the following patent applications: FKBP5: a novel target for antidepressant therapy. European Patent# EP 1687443 B1; Polymorphisms in ABCB1 associated with a lack of clinical response to medicaments. United States Patent # 8030033; Means and methods for diagnosing predisposition for treatment emergent suicidal ideation (TESI). European application number: 08016477.5 International application number: PCT/EP2009/061575. The remaining authors declare no competing interests.
Figures