Plasmid Localization and Partition in Enterobacteriaceae
- PMID: 31187729
- PMCID: PMC11573283
- DOI: 10.1128/ecosalplus.ESP-0003-2019
Plasmid Localization and Partition in Enterobacteriaceae
Abstract
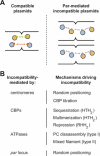
Plasmids are ubiquitous in the microbial world and have been identified in almost all species of bacteria that have been examined. Their localization inside the bacterial cell has been examined for about two decades; typically, they are not randomly distributed, and their positioning depends on copy number and their mode of segregation. Low-copy-number plasmids promote their own stable inheritance in their bacterial hosts by encoding active partition systems, which ensure that copies are positioned in both halves of a dividing cell. High-copy plasmids rely on passive diffusion of some copies, but many remain clustered together in the nucleoid-free regions of the cell. Here we review plasmid localization and partition (Par) systems, with particular emphasis on plasmids from Enterobacteriaceae and on recent results describing the in vivo localization properties and molecular mechanisms of each system. Partition systems also cause plasmid incompatibility such that distinct plasmids (with different replicons) with the same Par system cannot be stably maintained in the same cells. We discuss how partition-mediated incompatibility is a consequence of the partition mechanism.
Figures

Similar articles
-
Intracellular Positioning Systems Limit the Entropic Eviction of Secondary Replicons Toward the Nucleoid Edges in Bacterial Cells.J Mol Biol. 2020 Feb 7;432(3):745-761. doi: 10.1016/j.jmb.2019.11.027. Epub 2020 Jan 11. J Mol Biol. 2020. PMID: 31931015
-
Partition locus-based classification of selected plasmids in Klebsiella pneumoniae, Escherichia coli and Salmonella enterica spp.: an additional tool.J Microbiol Methods. 2015 Mar;110:85-91. doi: 10.1016/j.mimet.2015.01.019. Epub 2015 Jan 24. J Microbiol Methods. 2015. PMID: 25623509
-
Atypical low-copy number plasmid segregation systems, all in one?Plasmid. 2023 Jul;127:102694. doi: 10.1016/j.plasmid.2023.102694. Epub 2023 Jun 8. Plasmid. 2023. PMID: 37301314 Review.
-
Differential Localization and Functional Specialization of parS Centromere-Like Sites in repABC Replicons of Alphaproteobacteria.Appl Environ Microbiol. 2022 Apr 26;88(8):e0020722. doi: 10.1128/aem.00207-22. Epub 2022 Apr 7. Appl Environ Microbiol. 2022. PMID: 35389251 Free PMC article.
-
Mechanisms of Theta Plasmid Replication in Enterobacteria and Implications for Adaptation to Its Host.EcoSal Plus. 2020 Nov;9(1):10.1128/ecosalplus.ESP-0026-2019. doi: 10.1128/ecosalplus.ESP-0026-2019. EcoSal Plus. 2020. PMID: 33210586 Free PMC article. Review.
Cited by
-
IncFV plasmid pED208: Sequence analysis and evidence for translocation of maintenance/leading region proteins through diverse type IV secretion systems.Plasmid. 2022 Sep-Nov;123-124:102652. doi: 10.1016/j.plasmid.2022.102652. Epub 2022 Oct 11. Plasmid. 2022. PMID: 36228885 Free PMC article.
-
Genomic analysis of plasmid content in food isolates of E. coli strongly supports its role as a reservoir for the horizontal transfer of virulence and antibiotic resistance genes.Plasmid. 2022 Sep-Nov;123-124:102650. doi: 10.1016/j.plasmid.2022.102650. Epub 2022 Sep 18. Plasmid. 2022. PMID: 36130651 Free PMC article.
-
Three ParA Dimers Cooperatively Assemble on Type Ia Partition Promoters.Genes (Basel). 2021 Aug 28;12(9):1345. doi: 10.3390/genes12091345. Genes (Basel). 2021. PMID: 34573327 Free PMC article.
-
Plasmid co-infection: linking biological mechanisms to ecological and evolutionary dynamics.Philos Trans R Soc Lond B Biol Sci. 2022 Jan 17;377(1842):20200478. doi: 10.1098/rstb.2020.0478. Epub 2021 Nov 29. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 34839701 Free PMC article. Review.
-
Examination of Rickettsial Host Range for Shuttle Vectors Based on dnaA and parA Genes from the pRM Plasmid of Rickettsia monacensis.Appl Environ Microbiol. 2022 Apr 12;88(7):e0021022. doi: 10.1128/aem.00210-22. Epub 2022 Mar 24. Appl Environ Microbiol. 2022. PMID: 35323021 Free PMC article.
References
-
- Jacob F, Brenner S, Cuzin F. 1963. On the regulation of DNA replication in bacteria. Cold Spring Harb Symp Quant Biol 228:329–348. 10.1101/SQB.1963.028.01.048. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

