Meta-analysis of genome-wide association studies for loin muscle area and loin muscle depth in two Duroc pig populations
- PMID: 31188900
- PMCID: PMC6561594
- DOI: 10.1371/journal.pone.0218263
Meta-analysis of genome-wide association studies for loin muscle area and loin muscle depth in two Duroc pig populations
Abstract
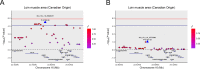
Loin muscle area (LMA) and loin muscle depth (LMD) are important traits influencing the production performance of breeding pigs. However, the genetic architecture of these two traits is still poorly understood. To discern the genetic architecture of LMA and LMD, a material consisting of 6043 Duroc pigs belonging to two populations with different genetic backgrounds was collected and applied in genome-wide association studies (GWAS) with a genome-wide distributed panel of 50K single nucleotide polymorphisms (SNPs). To improve the power of detection for common SNPs, we conducted a meta-analysis in these two pig populations and uncovered additional significant SNPs. As a result, we identified 75 significant SNPs for LMA and LMD on SSC6, 7, 12, 16, and 18. Among them, 25 common SNPs were associated with LMA and LMD. One pleiotropic quantitative trait locus (QTL), which was located on SSC7 with a 283 kb interval, was identified to affect LMA and LMD. Marker ALGA0040260 is a key SNP for this QTL, explained 1.77% and 2.48% of the phenotypic variance for LMA and LMD, respectively. Another genetic region on SSC16 (709 kb) was detected and displayed prominent association with LMA and the peak SNP, WU_10.2_16_35829257, contributed 1.83% of the phenotypic variance for LMA. Further bioinformatics analysis determined eight promising candidate genes (GCLC, GPX8, DAXX, FGF21, TAF11, SPDEF, NUDT3, and PACSIN1) with functions in glutathione metabolism, adipose and muscle tissues development and lipid metabolism. This study provides the first GWAS for the LMA and LMD of Duroc breed to analyze the underlying genetic variants through a large sample size. The findings further advance our understanding and help elucidate the genetic architecture of LMA, LMD and growth-related traits in pigs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

