The landscape of transcription initiation across latent and lytic KSHV genomes
- PMID: 31188901
- PMCID: PMC6590836
- DOI: 10.1371/journal.ppat.1007852
The landscape of transcription initiation across latent and lytic KSHV genomes
Abstract
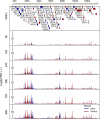
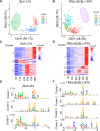
Precise promoter annotation is required for understanding the mechanistic basis of transcription initiation. In the context of complex genomes, such as herpesviruses where there is extensive genic overlap, identification of transcription start sites (TSSs) is particularly problematic and cannot be comprehensively accessed by standard RNA sequencing approaches. Kaposi's sarcoma-associated herpesvirus (KSHV) is an oncogenic gammaherpesvirus and the etiological agent of Kaposi's sarcoma and the B cell lymphoma primary effusion lymphoma (PEL). Here, we leverage RNA annotation and mapping of promoters for analysis of gene expression (RAMPAGE) and define KSHV TSSs transcriptome-wide and at nucleotide resolution in two widely used models of KSHV infection, namely iSLK.219 cells and the PEL cell line TREx-BCBL1-RTA. By mapping TSSs over a 96 h time course of reactivation we confirm 48 of 50 previously identified TSSs. Moreover, we identify over 100 novel transcription start site clusters (TSCs) in each cell line. Our analyses identified cell-type specific differences in TSC positions as well as promoter strength, and defined motifs within viral core promoters. Collectively, by defining TSSs at high resolution we have greatly expanded the transcriptional landscape of the KSHV genome and identified transcriptional control mechanisms at play during KSHV lytic reactivation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100(26):15776–81. Epub 2003/12/08. 10.1073/pnas.2136655100 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

