Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages
- PMID: 31189557
- PMCID: PMC6589867
- DOI: 10.1128/CMR.00135-18
Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages
Abstract
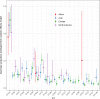
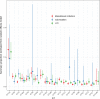
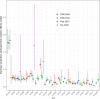
Extraintestinal pathogenic Escherichia coli (ExPEC) strains are responsible for a majority of human extraintestinal infections globally, resulting in enormous direct medical and social costs. ExPEC strains are comprised of many lineages, but only a subset is responsible for the vast majority of infections. Few systematic surveillance systems exist for ExPEC. To address this gap, we systematically reviewed and meta-analyzed 217 studies (1995 to 2018) that performed multilocus sequence typing or whole-genome sequencing to genotype E. coli recovered from extraintestinal infections or the gut. Twenty major ExPEC sequence types (STs) accounted for 85% of E. coli isolates from the included studies. ST131 was the most common ST from 2000 onwards, covering all geographic regions. Antimicrobial resistance-based isolate study inclusion criteria likely led to an overestimation and underestimation of some lineages. European and North American studies showed similar distributions of ExPEC STs, but Asian and African studies diverged. Epidemiology and population dynamics of ExPEC are complex; summary proportion for some STs varied over time (e.g., ST95), while other STs were constant (e.g., ST10). Persistence, adaptation, and predominance in the intestinal reservoir may drive ExPEC success. Systematic, unbiased tracking of predominant ExPEC lineages will direct research toward better treatment and prevention strategies for extraintestinal infections.
Keywords: Escherichia coli; extraintestinal infections; extraintestinal pathogenic E. coli; molecular epidemiology.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi:10.1016/S1473-3099(15)00424-7. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

