HIV Diversity and Genetic Compartmentalization in Blood and Testes during Suppressive Antiretroviral Therapy
- PMID: 31189714
- PMCID: PMC6694813
- DOI: 10.1128/JVI.00755-19
HIV Diversity and Genetic Compartmentalization in Blood and Testes during Suppressive Antiretroviral Therapy
Abstract
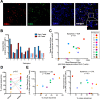
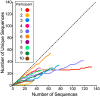
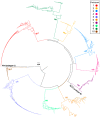
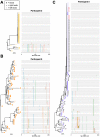
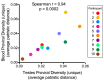
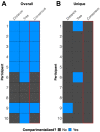
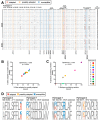
HIV's ability to persist during suppressive antiretroviral therapy is the main barrier to cure. Immune-privileged tissues, such as the testes, may constitute distinctive sites of HIV persistence, but this has been challenging to study in humans. We analyzed the proviral burden and genetics in the blood and testes of 10 individuals on suppressive therapy who underwent elective gender-affirming surgery. HIV DNA levels in matched blood and testes were quantified by quantitative PCR, and subgenomic proviral sequences (nef region) were characterized from single templates. HIV diversity, compartmentalization, and immune escape burden were assessed using genetic and phylogenetic approaches. Diverse proviruses were recovered from the blood (396 sequences; 354 nef-intact sequences) and testes (326 sequences; 309 nef-intact sequences) of all participants. Notably, the frequency of identical HIV sequences varied markedly between and within individuals. Nevertheless, proviral loads, within-host unique HIV sequence diversity, and the immune escape burden correlated positively between blood and testes. When all intact nef sequences were evaluated, 60% of participants exhibited significant blood-testis genetic compartmentalization, but none did so when the evaluation was restricted to unique sequences per site, suggesting that compartmentalization, when present, is attributable to the clonal expansion of HIV-infected cells. Our observations confirm the testes as a site of HIV persistence and suggest that individuals with larger and more diverse blood reservoirs will have larger and more diverse testis reservoirs. Furthermore, while the testis microenvironment may not be sufficiently unique to facilitate the seeding of unique viral populations therein, differential clonal expansion dynamics may be at play, which may complicate HIV eradication.IMPORTANCE Two key questions in HIV reservoir biology are whether immune-privileged tissues, such as the testes, harbor distinctive proviral populations during suppressive therapy and, if so, by what mechanism. While our results indicated that blood-testis HIV genetic compartmentalization was reasonably common (60%), it was always attributable to differential frequencies of identical HIV sequences between sites. No blood-tissue data set retained evidence of compartmentalization when only unique HIV sequences per site were considered; moreover, HIV immune escape mutation burdens were highly concordant between sites. We conclude that the principal mechanism by which blood and testis reservoirs differ is not via seeding of divergent HIV sequences therein but, rather, via differential clonal expansion of latently infected cells. Thus, while viral diversity and escape-related barriers to HIV eradication are of a broadly similar magnitude across the blood and testes, clonal expansion represents a challenge. The results support individualized analysis of within-host reservoir diversity to inform curative approaches.
Keywords: HIV; clonal expansion; diversity; genetic compartmentalization; reservoir; testes.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi:10.1126/science.278.5341.1295. - DOI - PubMed
-
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 73:10489–10502. - PMC - PubMed
-
- Herbeck JT, Rolland M, Liu Y, McLaughlin S, McNevin J, Zhao H, Wong K, Stoddard JN, Raugi D, Sorensen S, Genowati I, Birditt B, McKay A, Diem K, Maust BS, Deng W, Collier AC, Stekler JD, McElrath MJ, Mullins JI. 2011. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol 85:7523–7534. doi:10.1128/JVI.02697-10. - DOI - PMC - PubMed
-
- Dapp MJ, Kober KM, Chen L, Westfall DH, Wong K, Zhao H, Hall BM, Deng W, Sibley T, Ghorai S, Kim K, Chen N, McHugh S, Au L, Cohen M, Anastos K, Mullins JI. 2017. Patterns and rates of viral evolution in HIV-1 subtype B infected females and males. PLoS One 12:e0182443. doi:10.1371/journal.pone.0182443. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

