Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome
- PMID: 31189953
- PMCID: PMC6774351
- DOI: 10.1038/s41586-019-1295-z
Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome
Abstract
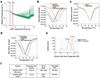
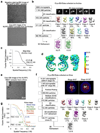
The NLRP3 inflammasome can be activated by stimuli that include nigericin, uric acid crystals, amyloid-β fibrils and extracellular ATP. The mitotic kinase NEK7 licenses the assembly and activation of the NLRP3 inflammasome in interphase. Here we report a cryo-electron microscopy structure of inactive human NLRP3 in complex with NEK7, at a resolution of 3.8 Å. The earring-shaped NLRP3 consists of curved leucine-rich-repeat and globular NACHT domains, and the C-terminal lobe of NEK7 nestles against both NLRP3 domains. Structural recognition between NLRP3 and NEK7 is confirmed by mutagenesis both in vitro and in cells. Modelling of an active NLRP3-NEK7 conformation based on the NLRC4 inflammasome predicts an additional contact between an NLRP3-bound NEK7 and a neighbouring NLRP3. Mutations to this interface abolish the ability of NEK7 or NLRP3 to rescue NLRP3 activation in NEK7-knockout or NLRP3-knockout cells. These data suggest that NEK7 bridges adjacent NLRP3 subunits with bipartite interactions to mediate the activation of the NLRP3 inflammasome.
Figures











Comment in
-
Enzymes that detoxify marine toxins.Nature. 2019 Jun;570(7761):315-316. doi: 10.1038/d41586-019-01742-1. Nature. 2019. PMID: 31213676 No abstract available.
-
A licence to kill during inflammation.Nature. 2019 Jun;570(7761):316-317. doi: 10.1038/d41586-019-01764-9. Nature. 2019. PMID: 31213677 No abstract available.
References
-
- Lamkanfi M & Dixit VM Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014). - PubMed
-
- Broz P & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16, 407–420 (2016). - PubMed
-
- Shi J, Gao W & Shao F Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem Sci (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

