Analysis of risk factors for colon cancer progression
- PMID: 31190895
- PMCID: PMC6535430
- DOI: 10.2147/OTT.S207390
Analysis of risk factors for colon cancer progression
Abstract
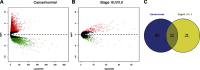
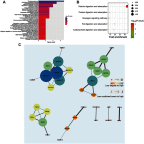
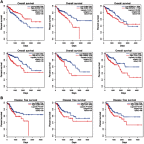
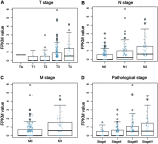
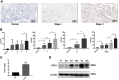
Purpose: This study aimed to find risk factors for colon cancer progression with bioinformatics methods, and validated by clinical patients. Methods: Differentially expressed genes (DEGs) between colon cancer tissues and normal colon tissues were extracted from The Cancer Genome Atlas (TCGA) database using R software, amounted to 8,051. DEGs between pathologic stage I+II and stage III+IV amounted to 373, and were compared with DEGs of cancer/normal analyzed above to get the intersection of both. Ninety-six intersected DEGs were identified and defined as progressive DEGs of colon cancer. Then these 96 progressive DEGs were studied by Gene ontology and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis using the DAVID database and visualizing by R software. A protein-protein interaction (PPI) network and functional modules were established using the STRING database. Further, an overall survival (OS) curve was drawn via the GEPIA website based on the CGA database and six progressive DEGs were found to be involved with OS of colon cancer patients. The Linkedomics website was used for detailed analysis of specific subsets of TNM. Results: Pregnancy specific glycoprotein (PSG), vitamin digestion, and absorption were confirmed to promote the progression of colon cancer. Furthermore, NTF4 was found to be associated with both OS and each subset of TNM; therefore, defined as a key risk factor for colon cancer progression. Further analysis of NTF4 expression using clinical data showed it acted as a key risk factor and diagnosis marker for colon cancer progression. Conclusion: NTF4 is a risk factor contributing to colon cancer progression and associated with overall survival.
Keywords: NTF4; colon cancer; differentially expressed genes; tumor progression.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures
References
-
- Yatsuoka T, Nishimura Y, Sakamoto H, Tanaka Y, Kurozumi M. [Lymph node metastasis of colorectal cancer with submucosal invasion]. Gan To Kagaku Ryoho. 2013;40(12):2041–2043. - PubMed
-
- Zhivotovskiy AS, Kutikhin AG, Azanov AZ, Yuzhalin AE, Magarill YA, Brusina EB. Colorectal cancer risk factors among the population of South-East Siberia: a case-control study. Asian Pac J Cancer Prev. 2012;13(10):5183–5188. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

