Dynamic Alterations in Yak Rumen Bacteria Community and Metabolome Characteristics in Response to Feed Type
- PMID: 31191470
- PMCID: PMC6538947
- DOI: 10.3389/fmicb.2019.01116
Dynamic Alterations in Yak Rumen Bacteria Community and Metabolome Characteristics in Response to Feed Type
Abstract
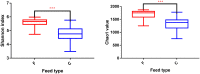
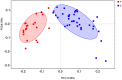
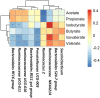
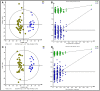
Current knowledge about the relationships between ruminal bacterial communities and metabolite profiles in the yak rumen is limited. This is due to differences in the nutritional and metabolic features between yak and other ordinary cattle combined with difficulties associated with farm-based research and a lack of technical guidance. A comprehensive analysis of the composition and alterations in ruminal metabolites is required to advance the development of modern yak husbandry. In the current study, we characterized the effect of feed type on the ruminal fluid microbiota and metabolites in yak using 16S rRNA gene sequencing and liquid chromatography-mass spectrometry (LC-MS). Bacteroidetes and Firmicutes were the predominant bacterial phyla in the yak rumen. At the genus level, the relative abundance of Bacteroidales BS11 gut group, Prevotellaceae UCG-003, Ruminococcaceae UCG-011, Bacteroidales RF16 group and Ruminococcaceae UCG-010 was significantly (P < 0.01) higher in the forage group compared to that in the concentrate group, while the concentrate group harbored higher proportions of Bacteroidales S24-7 group, Ruminococcaceae NK4A214, Succiniclasticum and Ruminococcus 2. Yak rumen metabolomics analysis combined with enrichment analysis revealed that feed type altered the concentrations of ruminal metabolites as well as the metabolic pattern, and significantly (P < 0.01) affected the concentrations of ruminal metabolites involved in protein digestion and absorption (e.g., L-arginine, ornithine, L-threonine, L-proline and β-alanine), purine metabolism (e.g., xanthine, hypoxanthine, deoxyadenosine and deoxyadenosine monophosphate) and fatty acid biosynthesis (e.g., stearic acid, myristic acid and arachidonic acid). Correlation analysis of the association of microorganisms with metabolite features provides us with a comprehensive understanding of the composition and function of microbial communities. Associations between utilization or production were widely identified between affected microbiota and certain metabolites, and these findings will contribute to the direction of future research in yak.
Keywords: feed type; metabolomics; microbiota; rumen; yak.
Figures




References
-
- Ametaj B. N., Zebeli Q., Saleem F., Psychogios N., Lewis M. J., Dunn S. M., et al. (2010). Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 6, 583–594. 10.1007/s11306-010-0227-6 - DOI
LinkOut - more resources
Full Text Sources

