Regulation of Gram-Positive Conjugation
- PMID: 31191478
- PMCID: PMC6540685
- DOI: 10.3389/fmicb.2019.01134
Regulation of Gram-Positive Conjugation
Abstract
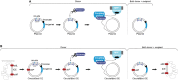
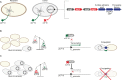
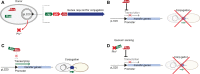
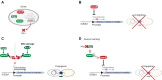
Type IV Secretion Systems (T4SSs) are membrane-spanning multiprotein complexes dedicated to protein secretion or conjugative DNA transport (conjugation systems) in bacteria. The prototype and best-characterized T4SS is that of the Gram-negative soil bacterium Agrobacterium tumefaciens. For Gram-positive bacteria, only conjugative T4SSs have been characterized in some biochemical, structural, and mechanistic details. These conjugation systems are predominantly encoded by self-transmissible plasmids but are also increasingly detected on integrative and conjugative elements (ICEs) and transposons. Here, we report regulatory details of conjugation systems from Enterococcus model plasmids pIP501 and pCF10, Bacillus plasmid pLS1, Clostridium plasmid pCW3, and staphylococcal plasmid pSK41. In addition, regulation of conjugative processes of ICEs (ICEBs1, ICESt1, ICESt3) by master regulators belonging to diverse repressor families will be discussed. A special focus of this review lies on the comparison of regulatory mechanisms executed by proteins belonging to the RRNPP family. These regulators share a common fold and govern several essential bacterial processes, including conjugative transfer.
Keywords: Gram-positive bacteria; conjugation system; integrative and conjugative element; plasmid; regulation; type IV secretion system.
Figures

Similar articles
-
Characterization of ConE, the VirB4 Homolog of the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2023 Jun 27;205(6):e0003323. doi: 10.1128/jb.00033-23. Epub 2023 May 23. J Bacteriol. 2023. PMID: 37219457 Free PMC article.
-
Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis.Plasmid. 2016 Jul;86:14-25. doi: 10.1016/j.plasmid.2016.07.001. Epub 2016 Jul 2. Plasmid. 2016. PMID: 27381852 Review.
-
Mechanisms of Conjugative Transfer and Type IV Secretion-Mediated Effector Transport in Gram-Positive Bacteria.Curr Top Microbiol Immunol. 2017;413:115-141. doi: 10.1007/978-3-319-75241-9_5. Curr Top Microbiol Immunol. 2017. PMID: 29536357
-
The 2.5 Å structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins.J Biol Chem. 2013 Jan 18;288(3):2018-28. doi: 10.1074/jbc.M112.428847. Epub 2012 Nov 27. J Biol Chem. 2013. PMID: 23188825 Free PMC article.
-
Conjugative type IV secretion systems in Gram-positive bacteria.Plasmid. 2013 Nov;70(3):289-302. doi: 10.1016/j.plasmid.2013.09.005. Epub 2013 Oct 12. Plasmid. 2013. PMID: 24129002 Free PMC article. Review.
Cited by
-
Selfish, promiscuous and sometimes useful: how mobile genetic elements drive horizontal gene transfer in microbial populations.Philos Trans R Soc Lond B Biol Sci. 2022 Oct 10;377(1861):20210234. doi: 10.1098/rstb.2021.0234. Epub 2022 Aug 22. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35989606 Free PMC article. Review.
-
Impact of Antibiotics as Waste, Physical, Chemical, and Enzymatical Degradation: Use of Laccases.Molecules. 2022 Jul 11;27(14):4436. doi: 10.3390/molecules27144436. Molecules. 2022. PMID: 35889311 Free PMC article. Review.
-
Microbial applications for sustainable space exploration beyond low Earth orbit.NPJ Microgravity. 2023 Jun 21;9(1):47. doi: 10.1038/s41526-023-00285-0. NPJ Microgravity. 2023. PMID: 37344487 Free PMC article. Review.
-
Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization.Nucleic Acids Res. 2020 Aug 20;48(14):8113-8127. doi: 10.1093/nar/gkaa540. Nucleic Acids Res. 2020. PMID: 32658272 Free PMC article.
-
New Platform for Screening Genetic Libraries at Elevated Temperatures: Biological and Genomic Information and Genetic Tools of Geobacillus thermodenitrificans K1041.Appl Environ Microbiol. 2022 Sep 22;88(18):e0105122. doi: 10.1128/aem.01051-22. Epub 2022 Sep 7. Appl Environ Microbiol. 2022. PMID: 36069579 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

