Inactivation of farR Causes High Rhodomyrtone Resistance and Increased Pathogenicity in Staphylococcus aureus
- PMID: 31191485
- PMCID: PMC6547885
- DOI: 10.3389/fmicb.2019.01157
Inactivation of farR Causes High Rhodomyrtone Resistance and Increased Pathogenicity in Staphylococcus aureus
Abstract
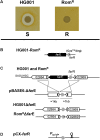
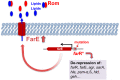
Rhodomyrtone (Rom) is an acylphloroglucinol antibiotic originally isolated from leaves of Rhodomyrtus tomentosa. Rom targets the bacterial membrane and is active against a wide range of Gram-positive bacteria but the exact mode of action remains obscure. Here we isolated and characterized a spontaneous Rom-resistant mutant from the model strain Staphylococcus aureus HG001 (RomR) to learn more about the resistance mechanism. We showed that Rom-resistance is based on a single point mutation in the coding region of farR [regulator of fatty acid (FA) resistance] that causes an amino acid change from Cys to Arg at position 116 in FarR, that affects FarR activity. Comparative transcriptome analysis revealed that mutated farR affects transcription of many genes in distinct pathways. FarR represses for example the expression of its own gene (farR), its flanking gene farE (effector of FA resistance), and other global regulators such as agr and sarA. All these genes were consequently upregulated in the RomR clone. Particularly the upregulation of agr and sarA leads to increased expression of virulence genes rendering the RomR clone more cytotoxic and more pathogenic in a mouse infection model. The Rom-resistance is largely due to the de-repression of farE. FarE is described as an efflux pump for linoleic and arachidonic acids. We observed an increased release of lipids in the RomR clone compared to its parental strain HG001. If farE is deleted in the RomR clone, or, if native farR is expressed in the RomR strain, the corresponding strains become hypersensitive to Rom. Overall, we show here that the high Rom-resistance is mediated by overexpression of farE in the RomR clone, that FarR is an important regulator, and that the point mutation in farR (RomR clone) makes the clone hyper-virulent.
Keywords: Gram-positive bacteria; Staphylococcus; antibiotic; membrane active; rhodomyrtone.
Figures


Similar articles
-
The Diverse Activities and Mechanisms of the Acylphloroglucinol Antibiotic Rhodomyrtone: Antibacterial Activity and Beyond.Antibiotics (Basel). 2024 Oct 2;13(10):936. doi: 10.3390/antibiotics13100936. Antibiotics (Basel). 2024. PMID: 39452203 Free PMC article. Review.
-
Molecular Basis of Rhodomyrtone Resistance in Staphylococcus aureus.mBio. 2021 Feb 22;13(1):e0383321. doi: 10.1128/mbio.03833-21. Epub 2022 Feb 15. mBio. 2021. PMID: 35164566 Free PMC article.
-
Inducible Expression of a Resistance-Nodulation-Division-Type Efflux Pump in Staphylococcus aureus Provides Resistance to Linoleic and Arachidonic Acids.J Bacteriol. 2015 Jun;197(11):1893-905. doi: 10.1128/JB.02607-14. Epub 2015 Mar 23. J Bacteriol. 2015. PMID: 25802299 Free PMC article.
-
DNA Binding and Sensor Specificity of FarR, a Novel TetR Family Regulator Required for Induction of the Fatty Acid Efflux Pump FarE in Staphylococcus aureus.J Bacteriol. 2019 Jan 11;201(3):e00602-18. doi: 10.1128/JB.00602-18. Print 2019 Feb 1. J Bacteriol. 2019. PMID: 30455282 Free PMC article.
-
[The role of cell wall organization and active efflux pump systems in multidrug resistance of bacteria].Mikrobiyol Bul. 2007 Apr;41(2):309-27. Mikrobiyol Bul. 2007. PMID: 17682720 Review. Turkish.
Cited by
-
The Diverse Activities and Mechanisms of the Acylphloroglucinol Antibiotic Rhodomyrtone: Antibacterial Activity and Beyond.Antibiotics (Basel). 2024 Oct 2;13(10):936. doi: 10.3390/antibiotics13100936. Antibiotics (Basel). 2024. PMID: 39452203 Free PMC article. Review.
-
Rhodomyrtus tomentosa Leaf Extract and Rhodomyrtone Combat Streptococcus pneumoniae Biofilm and Inhibit Invasiveness to Human Lung Epithelial and Enhance Pneumococcal Phagocytosis by Macrophage.Curr Microbiol. 2020 Nov;77(11):3546-3554. doi: 10.1007/s00284-020-02164-3. Epub 2020 Aug 18. Curr Microbiol. 2020. PMID: 32812080
-
Staphylococcus aureus Releases Proinflammatory Membrane Vesicles To Resist Antimicrobial Fatty Acids.mSphere. 2020 Sep 30;5(5):e00804-20. doi: 10.1128/mSphere.00804-20. mSphere. 2020. PMID: 32999082 Free PMC article.
-
Repeated Emergence of Variant TetR Family Regulator, FarR, and Increased Resistance to Antimicrobial Unsaturated Fatty Acid among Clonal Complex 5 Methicillin-Resistant Staphylococcus aureus.Antimicrob Agents Chemother. 2023 Mar 16;67(3):e0074922. doi: 10.1128/aac.00749-22. Epub 2023 Feb 6. Antimicrob Agents Chemother. 2023. PMID: 36744906 Free PMC article.
-
The identification of two M20B family peptidases required for full virulence in Staphylococcus aureus.Front Cell Infect Microbiol. 2023 Jul 19;13:1176769. doi: 10.3389/fcimb.2023.1176769. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37538308 Free PMC article.
References
-
- Alnaseri H., Arsic B., Schneider J. E., Kaiser J. C., Scinocca Z. C., Heinrichs D. E., et al. (2015). Inducible expression of a resistance-nodulation-division-type efflux pump in Staphylococcus aureus provides resistance to linoleic and arachidonic acids. J. Bacteriol. 197 1893–1905. 10.1128/JB.02607-2614 - DOI - PMC - PubMed
-
- Cherkaoui A., Diene S. M., Fischer A., Leo S., Francois P., Schrenzel J. (2017). Transcriptional modulation of penicillin-binding protein 1b, outer membrane protein P2 and efflux pump (AcrAB-TolC) during heat stress is correlated to enhanced bactericidal action of imipenem on non-typeable Haemophilus influenzae. Front. Microbiol. 8:2676. 10.3389/fmicb.2017.02676 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

