Differential Expression of Three Cryptosporidium Species-Specific MEDLE Proteins
- PMID: 31191495
- PMCID: PMC6549896
- DOI: 10.3389/fmicb.2019.01177
Differential Expression of Three Cryptosporidium Species-Specific MEDLE Proteins
Abstract
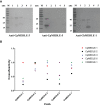
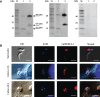
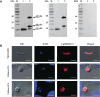
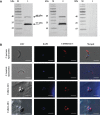
Cryptosporidium parvum and Cryptosporidium hominis share highly similar proteomes, with merely ~3% divergence in overall nucleotide sequences. Cryptosporidium-specific MEDLE family is one of the major differences in gene content between the two species. Comparative genomic analysis indicated that MEDLE family may contribute to differences in host range among Cryptosporidium spp. Previous studies have suggested that CpMEDLE-1 encoded by cgd5_4580 and CpMEDLE-2 encoded by cgd5_4590 are potentially involved in the invasion of C. parvum. In this study, we expressed in Escherichia coli, the C. hominis-specific member of the MEDLE protein family, ChMEDLE-1 encoded by chro.50507, and two C. parvum-specific members, CpMEDLE-3 encoded by cgd5_4600 and CpMEDLE-5 encoded by cgd6_5480. Quantitative PCR, immunofluorescence staining and in vitro neutralization assay were conducted to assess their biologic characteristics. The expression of the cgd5_4600 gene was high during 12-48 h of the in vitro culture, while the expression of cgd6_5480 was the highest at 2 h. ChMEDLE-1 and CpMEDLE-3 proteins were mostly located in the anterior and mid-anterior region of sporozoites and merozoites, whereas CpMEDLE-5 was expressed over the entire surface of these invasive stages. Polyclonal antibodies against MEDLE proteins had different neutralization efficiency, reaching approximately 50% for ChMEDLE-1 and 60% for CpMEDLE-3, but only 20% for CpMEDLE-5. The differences in protein and gene expression and neutralizing capacity indicated the MEDLE proteins may have different roles during Cryptosporidium invasion and growth.
Keywords: Cryptosporidium hominis; Cryptosporidium parvum; MEDLE family; growth; invasion.
Figures

References
-
- Cai X., Woods K. M., Upton S. J., Zhu G. (2005). Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrob. Agents Chemother. 49, 4437–4442. 10.1128/AAC.49.11.4437-4442.2005, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

