Functional, Structural and Biochemical Features of Plant Serinyl-Glutathione Transferases
- PMID: 31191562
- PMCID: PMC6540824
- DOI: 10.3389/fpls.2019.00608
Functional, Structural and Biochemical Features of Plant Serinyl-Glutathione Transferases
Abstract
Glutathione transferases (GSTs) belong to a ubiquitous multigenic family of enzymes involved in diverse biological processes including xenobiotic detoxification and secondary metabolism. A canonical GST is formed by two domains, the N-terminal one adopting a thioredoxin (TRX) fold and the C-terminal one an all-helical structure. The most recent genomic and phylogenetic analysis based on this domain organization allowed the classification of the GST family into 14 classes in terrestrial plants. These GSTs are further distinguished based on the presence of the ancestral cysteine (Cys-GSTs) present in TRX family proteins or on its substitution by a serine (Ser-GSTs). Cys-GSTs catalyze the reduction of dehydroascorbate and deglutathionylation reactions whereas Ser-GSTs catalyze glutathione conjugation reactions and eventually have peroxidase activity, both activities being important for stress tolerance or herbicide detoxification. Through non-catalytic, so-called ligandin properties, numerous plant GSTs also participate in the binding and transport of small heterocyclic ligands such as flavonoids including anthocyanins, and polyphenols. So far, this function has likely been underestimated compared to the other documented roles of GSTs. In this review, we compiled data concerning the known enzymatic and structural properties as well as the biochemical and physiological functions associated to plant GSTs having a conserved serine in their active site.
Keywords: glutathione transferases; ligandin property; photosynthetic organisms; phylogeny; secondary metabolism; structure; xenobiotic detoxification.
Figures


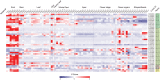
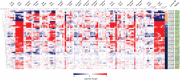
 . Residues that participate in dimer stabilization via strong polar interactions are in bold and marked with ∗. Residues involved in binding glutathione (G-site) are in bold type, highlighted yellow, and marked with
. Residues that participate in dimer stabilization via strong polar interactions are in bold and marked with ∗. Residues involved in binding glutathione (G-site) are in bold type, highlighted yellow, and marked with  . Residues of the characterized H-sites are in bold type, highlighted green, and marked with
. Residues of the characterized H-sites are in bold type, highlighted green, and marked with  . Residues of the L1-site (GmGSTU4, 2VO4) are in bold type, highlighted red, and marked with
. Residues of the L1-site (GmGSTU4, 2VO4) are in bold type, highlighted red, and marked with  . Residues of the L2-site (AtGSTF2, 5A4U, 5A4V, and 5A4W) are in bold type, highlighted blue, and marked with
. Residues of the L2-site (AtGSTF2, 5A4U, 5A4V, and 5A4W) are in bold type, highlighted blue, and marked with  . Residues of the L3-site (AtGSTF2, 5A4K, 5A4U, and 5A4W) are in bold type, highlighted pink, and marked with
. Residues of the L3-site (AtGSTF2, 5A4K, 5A4U, and 5A4W) are in bold type, highlighted pink, and marked with  .
.
Similar articles
-
The still mysterious roles of cysteine-containing glutathione transferases in plants.Front Pharmacol. 2014 Aug 20;5:192. doi: 10.3389/fphar.2014.00192. eCollection 2014. Front Pharmacol. 2014. PMID: 25191271 Free PMC article. Review.
-
Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs).Vitam Horm. 2005;72:155-202. doi: 10.1016/S0083-6729(05)72005-7. Vitam Horm. 2005. PMID: 16492471 Review.
-
Multiple roles for plant glutathione transferases in xenobiotic detoxification.Drug Metab Rev. 2011 May;43(2):266-80. doi: 10.3109/03602532.2011.552910. Epub 2011 Mar 22. Drug Metab Rev. 2011. PMID: 21425939 Review.
-
Plant glutathione transferases.Methods Enzymol. 2005;401:169-86. doi: 10.1016/S0076-6879(05)01011-6. Methods Enzymol. 2005. PMID: 16399386
-
Glutathione transferases: a structural perspective.Drug Metab Rev. 2011 May;43(2):138-51. doi: 10.3109/03602532.2011.558093. Epub 2011 Mar 23. Drug Metab Rev. 2011. PMID: 21428697 Review.
Cited by
-
Repurposing Glutathione Transferases: Directed Evolution Combined with Chemical Modification for the Creation of a Semisynthetic Enzyme with High Hydroperoxidase Activity.Antioxidants (Basel). 2023 Dec 25;13(1):41. doi: 10.3390/antiox13010041. Antioxidants (Basel). 2023. PMID: 38247466 Free PMC article.
-
First in vivo Evidence That Glutathione-S-Transferase Operates in Photo-Oxidative Stress in Cyanobacteria.Front Microbiol. 2019 Aug 13;10:1899. doi: 10.3389/fmicb.2019.01899. eCollection 2019. Front Microbiol. 2019. PMID: 31456794 Free PMC article.
-
Reactive Oxygen Species: A Crosslink between Plant and Human Eukaryotic Cell Systems.Int J Mol Sci. 2023 Aug 22;24(17):13052. doi: 10.3390/ijms241713052. Int J Mol Sci. 2023. PMID: 37685857 Free PMC article. Review.
-
Glutathione S-transferase in mediating adaptive responses of oats (Avena sativa) to osmotic and cadmium stress: genome-wide analysis.BMC Plant Biol. 2025 Apr 25;25(1):538. doi: 10.1186/s12870-025-06559-x. BMC Plant Biol. 2025. PMID: 40281415 Free PMC article.
-
Review of protein structure-based analyses that illuminate plant stress mechanisms.Comput Struct Biotechnol J. 2025 Jul 13;27:3155-3166. doi: 10.1016/j.csbj.2025.07.021. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40727428 Free PMC article. Review.
References
-
- Axarli I., Dhavala P., Papageorgiou A. C., Labrou N. E. (2009a). Crystallographic and functional characterization of the fluorodifen-inducible glutathione transferase from Glycine max reveals an active site topography suited for diphenylether herbicides and a novel L-site. J. Mol. Biol. 385 984–1002. 10.1016/j.jmb.2008.10.084 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

