Plant Aquaporins in Infection by and Immunity Against Pathogens - A Critical Review
- PMID: 31191567
- PMCID: PMC6546722
- DOI: 10.3389/fpls.2019.00632
Plant Aquaporins in Infection by and Immunity Against Pathogens - A Critical Review
Abstract
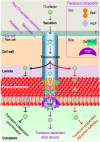
Plant aquaporins (AQPs) of the plasma membrane intrinsic protein (PIP) family face constant risk of hijack by pathogens aiming to infect plants. PIPs can also be involved in plant immunity against infection. This review will utilize two case studies to discuss biochemical and structural mechanisms that govern the functions of PIPs in the regulation of plant infection and immunity. The first example concerns the interaction between rice Oryza sativa and the bacterial blight pathogen Xanthomonas oryzae pv. oryzae (Xoo). To infect rice, Xoo uses the type III (T3) secretion system to secrete the proteic translocator Hpa1, and Hpa1 subsequently mediates the translocation of T3 effectors secreted by this system. Once shifted from bacteria into rice cells, effectors exert virulent or avirulent effects depending on the susceptibility of the rice varieties. The translocator function of Hpa1 requires cooperation with OsPIP1;3, the rice interactor of Hpa1. This role of OsPIP1;3 is related to regulatory models of effector translocation. The regulatory models have been proposed as, translocon-dependent delivery, translocon-independent pore formation, and effector endocytosis with membrane protein/lipid trafficking. The second case study includes the interaction of Hpa1 with the H2O2 transport channel AtPIP1;4, and the associated consequence for H2O2 signal transduction of immunity pathways in Arabidopsis thaliana, a non-host of Xoo. H2O2 is generated in the apoplast upon induction by a pathogen or microbial pattern. H2O2 from this source translocates quickly into Arabidopsis cells, where it interacts with pathways of intracellular immunity to confer plant resistance against diseases. To expedite H2O2 transport, AtPIP1;4 must adopt a specific conformation in a number of ways, including channel width extension through amino acid interactions and selectivity for H2O2 through amino acid protonation and tautomeric reactions. Both topics will reference relevant studies, conducted on other organisms and AQPs, to highlight possible mechanisms of T3 effector translocation currently under debate, and highlight the structural basis of AtPIP1;4 in H2O2 transport facilitated by gating and trafficking regulation.
Keywords: H2O2 transport; aquaporin; immunity signaling; plasma membrane intrinsic protein; translocon; type III effectors.
Figures


Similar articles
-
Hpa1 is a type III translocator in Xanthomonas oryzae pv. oryzae.BMC Microbiol. 2018 Sep 4;18(1):105. doi: 10.1186/s12866-018-1251-3. BMC Microbiol. 2018. PMID: 30180793 Free PMC article.
-
Functional modulation of an aquaporin to intensify photosynthesis and abrogate bacterial virulence in rice.Plant J. 2021 Oct;108(2):330-346. doi: 10.1111/tpj.15427. Epub 2021 Sep 18. Plant J. 2021. PMID: 34273211
-
Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways.Plant Physiol. 2016 Jul;171(3):1635-50. doi: 10.1104/pp.15.01237. Epub 2016 Mar 4. Plant Physiol. 2016. PMID: 26945050 Free PMC article.
-
Biological significance and topological basis of aquaporin-partnering protein-protein interactions.Plant Signal Behav. 2015;10(12):e1011947. doi: 10.1080/15592324.2015.1011947. Plant Signal Behav. 2015. PMID: 26786009 Free PMC article. Review.
-
Resistance Genes and their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)-an Updated Review.Rice (N Y). 2020 Jan 8;13(1):3. doi: 10.1186/s12284-019-0358-y. Rice (N Y). 2020. PMID: 31915945 Free PMC article. Review.
Cited by
-
High-Throughput Customization of Plant Microbiomes for Sustainable Agriculture.Front Plant Sci. 2020 Sep 8;11:569742. doi: 10.3389/fpls.2020.569742. eCollection 2020. Front Plant Sci. 2020. PMID: 33013992 Free PMC article. Review.
-
Prokaryotic Aquaporins.Cells. 2019 Oct 24;8(11):1316. doi: 10.3390/cells8111316. Cells. 2019. PMID: 31653102 Free PMC article. Review.
-
Rice aquaporin OsPIP2;2 is a water-transporting facilitator in relevance to drought-tolerant responses.Plant Direct. 2021 Aug 16;5(8):e338. doi: 10.1002/pld3.338. eCollection 2021 Aug. Plant Direct. 2021. PMID: 34430793 Free PMC article.
-
Cell-Wall-Degrading Enzymes-Related Genes Originating from Rhizoctonia solani Increase Sugar Beet Root Damage in the Presence of Leuconostoc mesenteroides.Int J Mol Sci. 2022 Jan 25;23(3):1366. doi: 10.3390/ijms23031366. Int J Mol Sci. 2022. PMID: 35163289 Free PMC article.
-
AtPIP1;4 and AtPIP2;4 Cooperatively Mediate H2O2 Transport to Regulate Plant Growth and Disease Resistance.Plants (Basel). 2024 Apr 3;13(7):1018. doi: 10.3390/plants13071018. Plants (Basel). 2024. PMID: 38611547 Free PMC article.
References
-
- Adam P. R., Barta M. L., Dickenson N. E. (2017). “Characterization of type three secretion system translocator interactions with phospholipid membranes,” in Type 3 Secretion Systems: Methods and Protocols, Methods in Molecular Biology, Vol. 1531 eds Nilles M. L., Condry D. L. J. (New York, NY: Springer Science + Business Media; ), 81–91. 10.1007/978-1-4939-6649-3_7 - DOI - PubMed
-
- Aguayo D., Pacheco N., Morales E. H., Collao B., Luraschi R., Cabezas C., et al. (2015). Hydrogen peroxide and hypochlorous acid influx through the major S. typhimurium porin OmpD is affected by substitution of key residues of the channel. Arch. Biochem. Biophys. 568 38–45. 10.1016/j.abb.2015.01.005 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

