A Quasi-Domesticate Relic Hybrid Population of Saccharomyces cerevisiae × S. paradoxus Adapted to Olive Brine
- PMID: 31191600
- PMCID: PMC6548830
- DOI: 10.3389/fgene.2019.00449
A Quasi-Domesticate Relic Hybrid Population of Saccharomyces cerevisiae × S. paradoxus Adapted to Olive Brine
Abstract
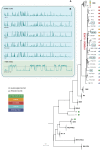
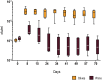
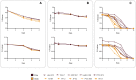
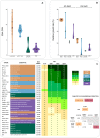
The adaptation of the yeast Saccharomyces cerevisiae to man-made environments for the fermentation of foodstuffs and beverages illustrates the scientific, social, and economic relevance of microbe domestication. Here we address a yet unexplored aspect of S. cerevisiae domestication, that of the emergence of lineages harboring some domestication signatures but that do not fit completely in the archetype of a domesticated yeast, by studying S. cerevisiae strains associated with processed olives, namely table olives, olive brine, olive oil, and alpechin. We confirmed earlier observations that reported that the Olives population results from a hybridization between S. cerevisiae and S. paradoxus. We concluded that the olive hybrids form a monophyletic lineage and that the S. cerevisiae progenitor belonged to the wine population of this species. We propose that homoploid hybridization gave rise to a diploid hybrid genome, which subsequently underwent the loss of most of the S. paradoxus sub-genome. Such a massive loss of heterozygosity was probably driven by adaptation to the new niche. We observed that olive strains are more fit to grow and survive in olive brine than control S. cerevisiae wine strains and that they appear to be adapted to cope with the presence of NaCl in olive brine through expansion of copy number of ENA genes. We also investigated whether the S. paradoxus HXT alleles retained by the Olives population were likely to contribute to the observed superior ability of these strains to consume sugars in brine. Our experiments indicate that sugar consumption profiles in the presence of NaCl are different between members of the Olives and Wine populations and only when cells are cultivated in nutritional conditions that support adaptation of their proteome to the high salt environment, which suggests that the observed differences are due to a better overall fitness of olives strains in the presence of high NaCl concentrations. Although relic olive hybrids exhibit several characteristics of a domesticated lineage, tangible benefits to humans cannot be associated with their phenotypes. These strains can be seen as a case of adaptation without positive or negative consequences to humans, that we define as a quasi-domestication.
Keywords: Saccharomyces cerevisiae; hybridization; microbe population genomics; microbiology of olive brine; yeast.
Figures

Similar articles
-
Brine salt concentration reduction and inoculation with autochthonous consortia: Impact on Protected Designation of Origin Nyons black table olive fermentations.Food Res Int. 2022 May;155:111069. doi: 10.1016/j.foodres.2022.111069. Epub 2022 Feb 28. Food Res Int. 2022. PMID: 35400447
-
Technological Improvement of Brined Black Table Olives Processed Using Two-Phase and Single-Phase Methods Under Slight CO2 Pressure and Low Salt Content.Foods. 2024 Nov 26;13(23):3799. doi: 10.3390/foods13233799. Foods. 2024. PMID: 39682870 Free PMC article.
-
Comparative genomics among Saccharomyces cerevisiae × Saccharomyces kudriavzevii natural hybrid strains isolated from wine and beer reveals different origins.BMC Genomics. 2012 Aug 20;13:407. doi: 10.1186/1471-2164-13-407. BMC Genomics. 2012. PMID: 22906207 Free PMC article.
-
The Ecology and Evolution of the Baker's Yeast Saccharomyces cerevisiae.Genes (Basel). 2022 Jan 26;13(2):230. doi: 10.3390/genes13020230. Genes (Basel). 2022. PMID: 35205274 Free PMC article. Review.
-
Yeast domestication.Curr Biol. 2025 Jun 9;35(11):R575-R586. doi: 10.1016/j.cub.2025.04.056. Curr Biol. 2025. PMID: 40494312 Review.
Cited by
-
The Rising Role of Omics and Meta-Omics in Table Olive Research.Foods. 2023 Oct 15;12(20):3783. doi: 10.3390/foods12203783. Foods. 2023. PMID: 37893676 Free PMC article. Review.
-
Hybridization and the origin of new yeast lineages.FEMS Yeast Res. 2020 Aug 1;20(5):foaa040. doi: 10.1093/femsyr/foaa040. FEMS Yeast Res. 2020. PMID: 32658267 Free PMC article. Review.
-
Mechanism of enhanced salt tolerance in Saccharomyces cerevisiae by CRZ1 overexpression.Sci Rep. 2024 Oct 2;14(1):22875. doi: 10.1038/s41598-024-74174-1. Sci Rep. 2024. PMID: 39358483 Free PMC article.
-
Multiple Hybridization Events Punctuate the Evolutionary Trajectory of Malassezia furfur.mBio. 2022 Apr 26;13(2):e0385321. doi: 10.1128/mbio.03853-21. Epub 2022 Apr 11. mBio. 2022. PMID: 35404119 Free PMC article.
-
A yeast living ancestor reveals the origin of genomic introgressions.Nature. 2020 Nov;587(7834):420-425. doi: 10.1038/s41586-020-2889-1. Epub 2020 Nov 11. Nature. 2020. PMID: 33177709
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

