Systems Chemical Genetics-Based Drug Discovery: Prioritizing Agents Targeting Multiple/Reliable Disease-Associated Genes as Drug Candidates
- PMID: 31191604
- PMCID: PMC6549477
- DOI: 10.3389/fgene.2019.00474
Systems Chemical Genetics-Based Drug Discovery: Prioritizing Agents Targeting Multiple/Reliable Disease-Associated Genes as Drug Candidates
Abstract
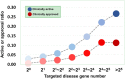
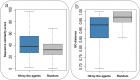
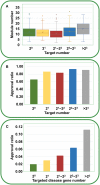
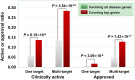
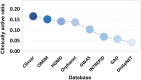
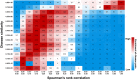
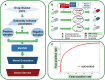
Genetic disease genes are considered a promising source of drug targets. Most diseases are caused by more than one pathogenic factor; thus, it is reasonable to consider that chemical agents targeting multiple disease genes are more likely to have desired activities. This is supported by a comprehensive analysis on the relationships between agent activity/druggability and target genetic characteristics. The therapeutic potential of agents increases steadily with increasing number of targeted disease genes, and can be further enhanced by strengthened genetic links between targets and diseases. By using the multi-label classification models for genetics-based drug activity prediction, we provide universal tools for prioritizing drug candidates. All of the documented data and the machine-learning prediction service are available at SCG-Drug (http://zhanglab.hzau.edu.cn/scgdrug).
Keywords: disease associated genes; drug discovery; drug targets; machine learning; systems chemical genetics.
Figures






References
LinkOut - more resources
Full Text Sources

