Downregulation of an Evolutionary Young miR-1290 in an iPSC-Derived Neural Stem Cell Model of Autism Spectrum Disorder
- PMID: 31191687
- PMCID: PMC6525818
- DOI: 10.1155/2019/8710180
Downregulation of an Evolutionary Young miR-1290 in an iPSC-Derived Neural Stem Cell Model of Autism Spectrum Disorder
Abstract
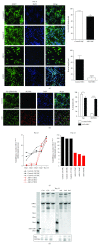
The identification of several evolutionary young miRNAs, which arose in primates, raised several possibilities for the role of such miRNAs in human-specific disease processes. We previously have identified an evolutionary young miRNA, miR-1290, to be essential in neural stem cell proliferation and neuronal differentiation. Here, we show that miR-1290 is significantly downregulated during neuronal differentiation in reprogrammed induced pluripotent stem cell- (iPSC-) derived neurons obtained from idiopathic autism spectrum disorder (ASD) patients. Further, we identified that miR-1290 is actively released into extracellular vesicles. Supplementing ASD patient-derived neural stem cells (NSCs) with conditioned media from differentiated control-NSCs spiked with "artificial EVs" containing synthetic miR-1290 oligonucleotides significantly rescued differentiation deficits in ASD cell lines. Based on our earlier published study and the observations from the data presented here, we conclude that miR-1290 regulation could play a critical role during neuronal differentiation in early brain development.
Figures



Similar articles
-
Profiling olfactory stem cells from living patients identifies miRNAs relevant for autism pathophysiology.Mol Autism. 2016 Jan 8;7:1. doi: 10.1186/s13229-015-0064-6. eCollection 2016. Mol Autism. 2016. PMID: 26753090 Free PMC article.
-
Stable expression of a truncated TLX variant drives differentiation of induced pluripotent stem cells into self-renewing neural stem cells for production of extracellular vesicles.Stem Cell Res Ther. 2022 Sep 2;13(1):436. doi: 10.1186/s13287-022-03131-4. Stem Cell Res Ther. 2022. PMID: 36056423 Free PMC article.
-
MicroRNA Profiling During Neural Differentiation of Induced Pluripotent Stem Cells.Int J Mol Sci. 2019 Jul 26;20(15):3651. doi: 10.3390/ijms20153651. Int J Mol Sci. 2019. PMID: 31357387 Free PMC article.
-
Optimizing neuronal differentiation from induced pluripotent stem cells to model ASD.Front Cell Neurosci. 2014 Apr 11;8:109. doi: 10.3389/fncel.2014.00109. eCollection 2014. Front Cell Neurosci. 2014. PMID: 24782713 Free PMC article. Review.
-
Understanding the molecular basis of autism in a dish using hiPSCs-derived neurons from ASD patients.Mol Brain. 2015 Sep 30;8(1):57. doi: 10.1186/s13041-015-0146-6. Mol Brain. 2015. PMID: 26419846 Free PMC article. Review.
Cited by
-
Systematic Review and Bioinformatic Analysis of microRNA Expression in Autism Spectrum Disorder Identifies Pathways Associated With Cancer, Metabolism, Cell Signaling, and Cell Adhesion.Front Psychiatry. 2021 Oct 21;12:630876. doi: 10.3389/fpsyt.2021.630876. eCollection 2021. Front Psychiatry. 2021. PMID: 34744804 Free PMC article.
-
A systematic review of non-coding RNA genes with differential expression profiles associated with autism spectrum disorders.PLoS One. 2023 Jun 15;18(6):e0287131. doi: 10.1371/journal.pone.0287131. eCollection 2023. PLoS One. 2023. PMID: 37319303 Free PMC article.
-
Signalling pathways in autism spectrum disorder: mechanisms and therapeutic implications.Signal Transduct Target Ther. 2022 Jul 11;7(1):229. doi: 10.1038/s41392-022-01081-0. Signal Transduct Target Ther. 2022. PMID: 35817793 Free PMC article. Review.
-
Methamphetamine Induces the Release of Proadhesive Extracellular Vesicles and Promotes Syncytia Formation: A Potential Role in HIV-1 Neuropathogenesis.Viruses. 2022 Mar 7;14(3):550. doi: 10.3390/v14030550. Viruses. 2022. PMID: 35336957 Free PMC article.
-
miRNAs as Predictors of Barrier Integrity.Biosensors (Basel). 2023 Mar 26;13(4):422. doi: 10.3390/bios13040422. Biosensors (Basel). 2023. PMID: 37185497 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

