Gene screening of colorectal cancers via network analysis
- PMID: 31191840
- PMCID: PMC6536014
Gene screening of colorectal cancers via network analysis
Abstract
Aim: Identifying crucial genes related to colorectal cancers via protein-protein interaction (PPI) network analysis is the aim of this study.
Background: colorectal cancer as major reason of mortality is evaluated by genetic and proteomic approaches to find suitable biomarkers. Chromosomal instability plays crucial role in CRC. Expression change of large numbers of genes is reported.
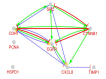
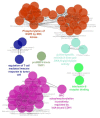
Methods: Differentially expressed genes related to CRCs which obtained from different proteomic methods were extracted from a review article of Paula Álvarez-Chaver et al. The genes interacted by Cytoscape software via STRING database. The central nodes determined and were enriched for biological terms by ClueGO. Action map for central genes was illustrated by CluePedia. The critical genes in CRC were introduced.
Results: Among 123 query genes, 114 one recognized by software and were included in the network. SRC, EGFR, PCNA, IL8, CTNNB1, TIMP1, CDH1, and HSPD1 were determined as central genes. After gene ontology analysis SRC, EGFR, and CDH1 were identified as critical genes related to CRC.
Conclusion: It seems that SRC, EGFR, and CDH1 and the related pathways are possible biomarkers for CRC.
Keywords: Biomarker; Colorectal cancer; Gene.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Evaluation of gene expression change in eosinophilic gastroenteritis.Gastroenterol Hepatol Bed Bench. 2019 Summer;12(3):239-245. Gastroenterol Hepatol Bed Bench. 2019. PMID: 31528308 Free PMC article.
-
Network analysis of grade II into grade III transition in rectum cancer patients.Gastroenterol Hepatol Bed Bench. 2018 Winter;11(Suppl 1):S118-S123. Gastroenterol Hepatol Bed Bench. 2018. PMID: 30774817 Free PMC article.
-
Unravelling Phyto-Compound Therapeutics against SRC Protein via FGF Pathway Targeting-A Comprehensive Approach Integrating Omics Data Analysis, Network Pharmacology, Virtual Screening, and Molecular Dynamics.Recent Adv Food Nutr Agric. 2024 Jun 25. doi: 10.2174/012772574X294492240527081926. Online ahead of print. Recent Adv Food Nutr Agric. 2024. PMID: 38919088
-
Identification of potential biomarkers with colorectal cancer based on bioinformatics analysis and machine learning.Math Biosci Eng. 2021 Oct 19;18(6):8997-9015. doi: 10.3934/mbe.2021443. Math Biosci Eng. 2021. PMID: 34814332
-
Assessment of the SRC Inhibition Role in the Efficacy of Breast Cancer Radiotherapy.J Lasers Med Sci. 2019 Fall;10(Suppl 1):S18-S22. doi: 10.15171/jlms.2019.S4. Epub 2019 Dec 1. J Lasers Med Sci. 2019. PMID: 32021668 Free PMC article. Review.
Cited by
-
Gene expression network analysis identified CDK1 and KIF11 as possible key molecules in the development of colorectal cancer from normal tissues.Genomics Inform. 2025 Jun 2;23(1):15. doi: 10.1186/s44342-025-00046-3. Genomics Inform. 2025. PMID: 40457491 Free PMC article.
-
14,15-Epoxyeicosatrienoic Acid Protect Against Glucose Deprivation and Reperfusion-Induced Cerebral Microvascular Endothelial Cells Injury by Modulating Mitochondrial Autophagy via SIRT1/FOXO3a Signaling Pathway and TSPO Protein.Front Cell Neurosci. 2022 Apr 26;16:888836. doi: 10.3389/fncel.2022.888836. eCollection 2022. Front Cell Neurosci. 2022. PMID: 35558879 Free PMC article.
-
Combinatorial Network of Transcriptional and miRNA Regulation in Colorectal Cancer.Int J Mol Sci. 2023 Mar 10;24(6):5356. doi: 10.3390/ijms24065356. Int J Mol Sci. 2023. PMID: 36982429 Free PMC article.
-
Assessment of colon cancer molecular mechanism: a system biology approach.Gastroenterol Hepatol Bed Bench. 2021 Fall;14(Suppl1):S51-S57. Gastroenterol Hepatol Bed Bench. 2021. PMID: 35154602 Free PMC article.
-
The effect of intestinal microbiota metabolites on HT29 cell line using MTT method in patients with colorectal cancer.Gastroenterol Hepatol Bed Bench. 2019;12(Suppl1):S74-S79. Gastroenterol Hepatol Bed Bench. 2019. PMID: 32099605 Free PMC article.
References
-
- Winawer SJ, Fletcher RH, Miller L, Godlee F, Stolar M, Mulrow C, et al. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterol. 1997;112:594–642. - PubMed
-
- Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–61. - PubMed
-
- Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–7. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
