Leveraging synthetic biology for producing bioactive polyketides and non-ribosomal peptides in bacterial heterologous hosts
- PMID: 31191858
- PMCID: PMC6540960
- DOI: 10.1039/c9md00055k
Leveraging synthetic biology for producing bioactive polyketides and non-ribosomal peptides in bacterial heterologous hosts
Abstract
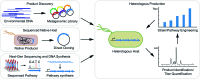
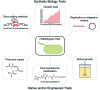
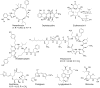
Bacteria have historically been a rich source of natural products (e.g. polyketides and non-ribosomal peptides) that possess medically-relevant activities. Despite extensive discovery programs in both industry and academia, a plethora of biosynthetic pathways remain uncharacterized and the corresponding molecular products untested for potential bioactivities. This knowledge gap comes in part from the fact that many putative natural product producers have not been cultured in conventional laboratory settings in which the corresponding products are produced at detectable levels. Next-generation sequencing technologies are further increasing the knowledge gap by obtaining metagenomic sequence information from complex communities where production of the desired compound cannot be isolated in the laboratory. For these reasons, many groups are turning to synthetic biology to produce putative natural products in heterologous hosts. This strategy depends on the ability to heterologously express putative biosynthetic gene clusters and produce relevant quantities of the corresponding products. Actinobacteria remain the most abundant source of natural products and the most promising heterologous hosts for natural product discovery and production. However, researchers are discovering more natural products from other groups of bacteria, such as myxobacteria and cyanobacteria. Therefore, phylogenetically similar heterologous hosts have become promising candidates for synthesizing these novel molecules. The downside of working with these microbes is the lack of well-characterized genetic tools for optimizing expression of gene clusters and product titers. This review examines heterologous expression of natural product gene clusters in terms of the motivations for this research, the traits desired in an ideal host, tools available to the field, and a survey of recent progress.
Figures

Similar articles
-
Stepwise genetic engineering of Pseudomonas putida enables robust heterologous production of prodigiosin and glidobactin A.Metab Eng. 2021 Sep;67:112-124. doi: 10.1016/j.ymben.2021.06.004. Epub 2021 Jun 24. Metab Eng. 2021. PMID: 34175462 Free PMC article.
-
Challenges and opportunities for natural product discovery, production, and engineering in native producers versus heterologous hosts.J Ind Microbiol Biotechnol. 2019 Mar;46(3-4):433-444. doi: 10.1007/s10295-018-2094-5. Epub 2018 Nov 13. J Ind Microbiol Biotechnol. 2019. PMID: 30426283 Free PMC article. Review.
-
Cell-free synthetic biology for natural product biosynthesis and discovery.Chem Soc Rev. 2025 May 6;54(9):4314-4352. doi: 10.1039/d4cs01198h. Chem Soc Rev. 2025. PMID: 40104998 Free PMC article. Review.
-
Discovery, characterization, and engineering of an advantageous Streptomyces host for heterologous expression of natural product biosynthetic gene clusters.Microb Cell Fact. 2024 May 24;23(1):149. doi: 10.1186/s12934-024-02416-y. Microb Cell Fact. 2024. PMID: 38790014 Free PMC article.
-
Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products.Fungal Genet Biol. 2016 Apr;89:18-28. doi: 10.1016/j.fgb.2016.01.012. Epub 2016 Jan 22. Fungal Genet Biol. 2016. PMID: 26808821 Free PMC article.
Cited by
-
Expression of fungal biosynthetic gene clusters in S. cerevisiae for natural product discovery.Synth Syst Biotechnol. 2021 Jan 25;6(1):20-22. doi: 10.1016/j.synbio.2021.01.003. eCollection 2021 Mar. Synth Syst Biotechnol. 2021. PMID: 33553706 Free PMC article.
-
Characterization of cross-species transcription and splicing from Penicillium to Saccharomyces cerevisiae.J Ind Microbiol Biotechnol. 2021 Dec 23;48(9-10):kuab054. doi: 10.1093/jimb/kuab054. J Ind Microbiol Biotechnol. 2021. PMID: 34387324 Free PMC article.
-
Expression of blue pigment synthetase a from Streptomyces lavenduale reveals insights on the effects of refactoring biosynthetic megasynthases for heterologous expression in Escherichia coli.Protein Expr Purif. 2023 Oct;210:106317. doi: 10.1016/j.pep.2023.106317. Epub 2023 Jun 5. Protein Expr Purif. 2023. PMID: 37286066 Free PMC article.
-
Bioprospecting Through Cloning of Whole Natural Product Biosynthetic Gene Clusters.Front Bioeng Biotechnol. 2020 Jun 5;8:526. doi: 10.3389/fbioe.2020.00526. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32582659 Free PMC article. Review.
-
The cyclic peptide G4CP2 enables the modulation of galactose metabolism in yeast by interfering with GAL4 transcriptional activity.Front Mol Biosci. 2023 Mar 1;10:1017757. doi: 10.3389/fmolb.2023.1017757. eCollection 2023. Front Mol Biosci. 2023. PMID: 36936986 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

