De novo production of aromatic m-cresol in Saccharomyces cerevisiae mediated by heterologous polyketide synthases combined with a 6-methylsalicylic acid decarboxylase
- PMID: 31193192
- PMCID: PMC6520567
- DOI: 10.1016/j.mec.2019.e00093
De novo production of aromatic m-cresol in Saccharomyces cerevisiae mediated by heterologous polyketide synthases combined with a 6-methylsalicylic acid decarboxylase
Abstract
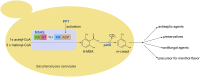
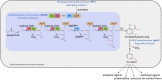
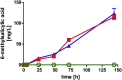
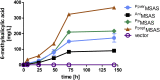
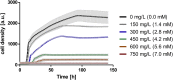
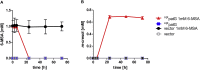
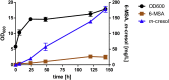
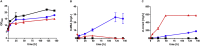
As a flavor and platform chemical, m-cresol (3-methylphenol) is a valuable industrial compound that currently is mainly synthesized by chemical methods from fossil resources. In this study, we present the first biotechnological de novo production of m-cresol from sugar in complex yeast extract-peptone medium with the yeast Saccharomyces cerevisiae. A heterologous pathway based on the decarboxylation of the polyketide 6-methylsalicylic acid (6-MSA) was introduced into a CEN.PK yeast strain. For synthesis of 6-MSA, expression of different variants of 6-MSA synthases (MSASs) were compared. Overexpression of codon-optimized MSAS from Penicillium patulum together with activating phosphopantetheinyl transferase npgA from Aspergillus nidulans resulted in up to 367 mg/L 6-MSA production. Additional genomic integration of the genes had a strongly promoting effect and 6-MSA titers reached more than 2 g/L. Simultaneous expression of 6-MSA decarboxylase patG from A. clavatus led to the complete conversion of 6-MSA and production of up to 589 mg/L m-cresol. As addition of 450-750 mg/L m-cresol to yeast cultures nearly completely inhibited growth our data suggest that the toxicity of m-cresol might be the limiting factor for higher production titers.
Keywords: 6-Methylsalicylic acid decarboxylase; 6-Methylsalicylic acid synthase; 6-methylsalicylic acid decarboxylase, PatG; 6-methylsalicylic acid synthase, MSAS; 6-methylsalicylic acid, 6-MSA; Acyl carrier protein, ACP; Acyltransferase, AT; Codon-optimization; Polyketide synthase; Saccharomyces cerevisiae; ketoreductase, KR; ketosynthase, KS; m-Cresol; optical density, OD; phosphopantetheinyl transferase, PPT; polyketide synthase, PKS; thioester hydrolase, TH.
Figures
Similar articles
-
The gene PatG involved in the biosynthesis pathway of patulin, a food-borne mycotoxin, encodes a 6-methylsalicylic acid decarboxylase.Int J Food Microbiol. 2014 Feb 3;171:77-83. doi: 10.1016/j.ijfoodmicro.2013.11.020. Epub 2013 Nov 27. Int J Food Microbiol. 2014. PMID: 24334092
-
De novo biosynthesis and gram-level production of m-cresol in Aspergillus nidulans.Appl Microbiol Biotechnol. 2021 Aug;105(16-17):6333-6343. doi: 10.1007/s00253-021-11490-w. Epub 2021 Aug 23. Appl Microbiol Biotechnol. 2021. PMID: 34423409
-
Improving 3-methylphenol (m-cresol) production in yeast via in vivo glycosylation or methylation.FEMS Yeast Res. 2020 Dec 16;20(8):foaa063. doi: 10.1093/femsyr/foaa063. FEMS Yeast Res. 2020. PMID: 33330906
-
Functional analysis of fungal polyketide biosynthesis genes.J Antibiot (Tokyo). 2010 May;63(5):207-18. doi: 10.1038/ja.2010.17. Epub 2010 Mar 5. J Antibiot (Tokyo). 2010. PMID: 20203700 Review.
-
Genes for polyketide secondary metabolic pathways in microorganisms and plants.Ciba Found Symp. 1992;171:88-106; discussion 106-12. doi: 10.1002/9780470514344.ch6. Ciba Found Symp. 1992. PMID: 1302187 Review.
Cited by
-
Recent advances in genetic engineering and chemical production in yeast species.FEMS Yeast Res. 2025 Jan 30;25:foaf009. doi: 10.1093/femsyr/foaf009. FEMS Yeast Res. 2025. PMID: 40082732 Free PMC article. Review.
-
Chaperone overexpression boosts heterologous small molecule production in Saccharomyces cerevisiae.Microb Cell Fact. 2025 May 19;24(1):112. doi: 10.1186/s12934-025-02728-7. Microb Cell Fact. 2025. PMID: 40383781 Free PMC article.
-
Novel bioreactor internals for the cultivation of spore-forming fungi in pellet form.Eng Life Sci. 2022 May 18;22(7):474-483. doi: 10.1002/elsc.202100094. eCollection 2022 Jul. Eng Life Sci. 2022. PMID: 35865648 Free PMC article.
-
Biosynthetic Potential of the Endophytic Fungus Helotiales sp. BL73 Revealed via Compound Identification and Genome Mining.Appl Environ Microbiol. 2022 Mar 22;88(6):e0251021. doi: 10.1128/aem.02510-21. Epub 2022 Feb 2. Appl Environ Microbiol. 2022. PMID: 35108081 Free PMC article.
-
First-class - biosynthesis of 6-MSA and bostrycoidin type I polyketides in Yarrowia lipolytica.Front Fungal Biol. 2024 Mar 22;5:1327777. doi: 10.3389/ffunb.2024.1327777. eCollection 2024. Front Fungal Biol. 2024. PMID: 38586602 Free PMC article.
References
-
- Beck J., Ripka S., Siegner A., Schiltz E., Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Eur. J. Biochem. 1990;192:487–498. - PubMed
-
- Berger R.G. Springer Berlin Heidelberg; 2007. Flavours and Fragrances Chemistry, Bioprocessing and Sustainability.
-
- Bruder S., Reifenrath M., Thomik T., Boles E., Herzog K. Parallelised online biomass monitoring in shake flasks enables efficient strain and carbon source dependent growth characterisation of Saccharomyces cerevisiae. Microb. Cell Factories. 2016;15:127. doi: 10.1186/s12934-016-0526-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

