C/D box snoRNAs in viral infections: RNA viruses use old dogs for new tricks
- PMID: 31193534
- PMCID: PMC6533054
- DOI: 10.1016/j.ncrna.2019.02.001
C/D box snoRNAs in viral infections: RNA viruses use old dogs for new tricks
Erratum in
-
Erratum regarding previously published articles.Noncoding RNA Res. 2020 Nov 7;5(4):220-221. doi: 10.1016/j.ncrna.2020.11.002. eCollection 2020 Dec. Noncoding RNA Res. 2020. PMID: 33294748 Free PMC article.
Abstract
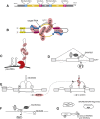
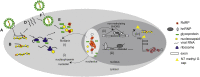
C/D box snoRNAs (SNORDs) are a highly expressed class of non-coding RNAs. Besides their well-established role in rRNA modification, C/D box snoRNAs form protein complexes devoid of fibrillarin and regulate pre-mRNA splicing and polyadenylation of numerous genes. There is an emerging body of evidence for functional interactions between RNA viruses and C/D box snoRNAs. The infectivity of some RNA viruses depends on enzymatically active fibrillarin, and many RNA viral proteins associate with nucleolin or nucleophosmin, suggesting that viruses benefit from their cytosolic accumulation. These interactions are likely reflected by morphological changes in the nucleolus, often leading to relocalization of nucleolar proteins and ncRNAs to the cytosol that are a characteristic feature of viral infections. Knock-down studies have also shown that RNA viruses need specific C/D box snoRNAs for optimal replication, suggesting that RNA viruses benefit from gene expression programs regulated by SNORDs, or that viruses have evolved "new" uses for these humble ncRNAs to advance their prospects during infection.
Figures
References
-
- Reddy R., Henning D., Busch H. Nucleotide sequence of nucleolar U3B RNA. J. Biol. Chem. 1979;254:11097–11105. - PubMed
-
- Maxwell E.S., Fournier M.J. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. - PubMed
-
- Shapiro E., Biezuner T., Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. - PubMed
-
- Bratkovic T., Rogelj B. The many faces of small nucleolar RNAs. Biochim. Biophys. Acta. 2014;1839:438–443. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

