qPCR data analysis: Better results through iconoclasm
- PMID: 31194178
- PMCID: PMC6554483
- DOI: 10.1016/j.bdq.2019.100084
qPCR data analysis: Better results through iconoclasm
Abstract
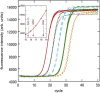
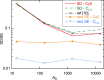
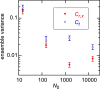
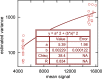
The standard approach for quantitative estimation of genetic materials with qPCR is calibration with known concentrations for the target substance, in which estimates of the quantification cycle (Cq ) are fitted to a straight-line function of log(N 0), where N 0 is the initial number of target molecules. The location of Cq for the unknown on this line then yields its N 0. The most widely used definition for Cq is an absolute threshold that falls in the early growth cycles. This usage is flawed as commonly implemented: threshold set very close to the baseline level, which is estimated separately, from designated "baseline cycles." The absolute threshold is especially poor for dealing with the scale variability often observed for growth profiles. Scale-independent markers, like the first derivative maximum (FDM) and a relative threshold (Cr ) avoid this problem. We describe improved methods for estimating these and other Cq markers and their standard errors, from a nonlinear algorithm that fits growth profiles to a 4-parameter log-logistic function plus a baseline function. Further, by examining six multidilution, multireplicate qPCR data sets, we find that nonlinear expressions are often preferred statistically for the dependence of Cq on log(N 0). This means that the amplification efficiency E depends on N 0, in violation of another tenet of qPCR analysis. Neglect of calibration nonlinearity leads to biased estimates of the unknown. By logic, E estimates from calibration fitting pertain to the earliest baseline cycles, not the early growth cycles used to estimate E from growth profiles for single reactions. This raises concern about the use of the latter in lengthy extrapolations to estimate N 0. Finally, we observe that replicate ensemble standard deviations greatly exceed predictions, implying that much better results can be achieved from qPCR through better experimental procedures, which likely include reducing pipette volume uncertainty.
Keywords: Calibration; Chi-square; Cq, quantification cycle; Ct, threshold cycle, where y = yq; Cy0, intersection of a straight line tangent to the curve at the FDM with the baseline-corrected x-axis; Data analysis; E, amplification efficiency; FDM and SDM, cycles where y reaches its maximal first and second derivatives, respectively; LS, least squares; N0, initial number of target molecules in sample; S, sum of weighted, squared residuals (= "Chisq" in KaleidaGraph fit results, = Χ2 when wi = 1/σi2); SD, standard deviation; SE, parameter standard error; Statistical errors; Weighted least squares; qPCR; qPCR, quantitative polymerase chain reaction; wi, statistical weight for ith data point; y and y0, fluorescence signal above baseline at cycle x and at cycle 0; yq, signal at x = Cq; Χ2, chi-square; ν, statistical degrees of freedom, = # of data points - # of adjustable parameters; σ2a and σ, variance and standard deviation.
Figures







Similar articles
-
Estimating Real-Time qPCR Amplification Efficiency from Single-Reaction Data.Life (Basel). 2021 Jul 14;11(7):693. doi: 10.3390/life11070693. Life (Basel). 2021. PMID: 34357065 Free PMC article.
-
Statistical uncertainty and its propagation in the analysis of quantitative polymerase chain reaction data: comparison of methods.Anal Biochem. 2014 Nov 1;464:94-102. doi: 10.1016/j.ab.2014.06.015. Epub 2014 Jun 30. Anal Biochem. 2014. PMID: 24991688
-
Absolute copy number from the statistics of the quantification cycle in replicate quantitative polymerase chain reaction experiments.Anal Chem. 2015 Feb 3;87(3):1889-95. doi: 10.1021/acs.analchem.5b00077. Epub 2015 Jan 23. Anal Chem. 2015. PMID: 25582662
-
Comparing real-time quantitative polymerase chain reaction analysis methods for precision, linearity, and accuracy of estimating amplification efficiency.Anal Biochem. 2014 Mar 15;449:76-82. doi: 10.1016/j.ab.2013.12.020. Epub 2013 Dec 21. Anal Biochem. 2014. PMID: 24365068
-
Use and Misuse of Cq in qPCR Data Analysis and Reporting.Life (Basel). 2021 May 29;11(6):496. doi: 10.3390/life11060496. Life (Basel). 2021. PMID: 34072308 Free PMC article. Review.
Cited by
-
Estimating Real-Time qPCR Amplification Efficiency from Single-Reaction Data.Life (Basel). 2021 Jul 14;11(7):693. doi: 10.3390/life11070693. Life (Basel). 2021. PMID: 34357065 Free PMC article.
-
Realizing the value in "non-standard" parts of the qPCR standard curve by integrating fundamentals of quantitative microbiology.Front Microbiol. 2023 Mar 3;14:1048661. doi: 10.3389/fmicb.2023.1048661. eCollection 2023. Front Microbiol. 2023. PMID: 36937263 Free PMC article.
-
Quantitative PCR of Small Nucleic Acids: Size Matters.ChemistrySelect. 2021 Mar 26;6(12):2975-2979. doi: 10.1002/slct.202100807. ChemistrySelect. 2021. PMID: 36819227 Free PMC article.
-
MicroRNA biomarkers as next-generation diagnostic tools for neurodegenerative diseases: a comprehensive review.Front Mol Neurosci. 2024 May 31;17:1386735. doi: 10.3389/fnmol.2024.1386735. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38883980 Free PMC article. Review.
-
Critique of the pairwise method for estimating qPCR amplification efficiency: beware of correlated data!BMC Bioinformatics. 2020 Jul 8;21(1):291. doi: 10.1186/s12859-020-03604-4. BMC Bioinformatics. 2020. PMID: 32640980 Free PMC article.
References
-
- Tellinghuisen J. Using nonlinear least squares to assess relative expression and its uncertainty in real-time qPCR studies. Anal. Biochem. 2016;496:1–3. - PubMed
-
- Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
