Identification of a 5-Gene Signature Predicting Progression and Prognosis of Clear Cell Renal Cell Carcinoma
- PMID: 31194719
- PMCID: PMC6587650
- DOI: 10.12659/MSM.917399
Identification of a 5-Gene Signature Predicting Progression and Prognosis of Clear Cell Renal Cell Carcinoma
Abstract
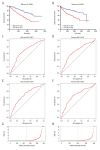
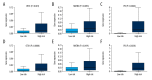
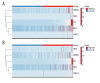
BACKGROUND Although the mortality rates of clear cell renal cell carcinoma (ccRCC) have decreased in recent years, the clinical outcome remains highly dependent on the individual patient. Therefore, identifying novel biomarkers for ccRCC patients is crucial. MATERIAL AND METHODS In this study, we obtained RNA sequencing data and clinical information from the TCGA database. Subsequently, we performed integrated bioinformatic analysis that includes differently expressed genes analysis, gene ontology and KEGG pathway analysis, protein-protein interaction analysis, and survival analysis. Moreover, univariate and multivariate Cox proportional hazards regression models were constructed. RESULTS As a result, we identified a total of 263 dysregulated genes that may participate in the metastasis of ccRCC, and established a predictive signature relying on the expression of OTX1, MATN4, PI3, ERVV-2, and NFE4, which could serve as significant progressive and prognostic biomarkers for ccRCC. CONCLUSIONS We identified differentially expressed genes that may be involved in the metastasis of ccRCC. Moreover, a predictive signature based on the expression of OTX1, MATN4, PI3, ERVV-2, and NFE4 could be an independent prognostic factor for ccRCC.
Conflict of interest statement
None.
Figures
Similar articles
-
A novel 10 glycolysis-related genes signature could predict overall survival for clear cell renal cell carcinoma.BMC Cancer. 2021 Apr 9;21(1):381. doi: 10.1186/s12885-021-08111-0. BMC Cancer. 2021. PMID: 33836688 Free PMC article.
-
A four-gene signature predicts survival in clear-cell renal-cell carcinoma.Oncotarget. 2016 Dec 13;7(50):82712-82726. doi: 10.18632/oncotarget.12631. Oncotarget. 2016. PMID: 27779101 Free PMC article.
-
Overexpression of Apolipoprotein C1 (APOC1) in Clear Cell Renal Cell Carcinoma and Its Prognostic Significance.Med Sci Monit. 2021 Feb 16;27:e929347. doi: 10.12659/MSM.929347. Med Sci Monit. 2021. PMID: 33591959 Free PMC article.
-
Identification of Prognostic Biomarkers for Clear Cell Renal Cell Carcinoma (ccRCC) by Transcriptomics.Ann Clin Lab Sci. 2021 Sep;51(5):597-608. Ann Clin Lab Sci. 2021. PMID: 34686501
-
Comprehensive analysis of a novel lncRNA profile reveals potential prognostic biomarkers in clear cell renal cell carcinoma.Oncol Rep. 2018 Sep;40(3):1503-1514. doi: 10.3892/or.2018.6540. Epub 2018 Jul 2. Oncol Rep. 2018. PMID: 30015930
Cited by
-
Two-Dimensional-PAGE Coupled with nLC-MS/MS-Based Identification of Differentially Expressed Proteins and Tumorigenic Pathways in MCF7 Breast Cancer Cells Transfected for JTB Protein Silencing.Molecules. 2023 Nov 9;28(22):7501. doi: 10.3390/molecules28227501. Molecules. 2023. PMID: 38005222 Free PMC article.
-
ADCY4 promotes brain metastasis in small cell lung cancer and is associated with energy metabolism.Heliyon. 2024 Mar 28;10(7):e28162. doi: 10.1016/j.heliyon.2024.e28162. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38596032 Free PMC article.
-
DNA Methylation-Based Panel Predicts Survival of Patients With Clear Cell Renal Cell Carcinoma and Its Correlations With Genomic Metrics and Tumor Immune Cell Infiltration.Front Cell Dev Biol. 2020 Oct 15;8:572628. doi: 10.3389/fcell.2020.572628. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33178689 Free PMC article.
-
Construction and validation of a prognostic model for kidney renal clear cell carcinoma based on podocyte-associated genes.Cancer Med. 2022 Oct;11(19):3549-3562. doi: 10.1002/cam4.4733. Epub 2022 Apr 4. Cancer Med. 2022. PMID: 35373928 Free PMC article.
-
Weighted Gene Coexpression Network Analysis Identifies TBC1D10C as a New Prognostic Biomarker for Breast Cancer.Anal Cell Pathol (Amst). 2022 Apr 5;2022:5259187. doi: 10.1155/2022/5259187. eCollection 2022. Anal Cell Pathol (Amst). 2022. PMID: 35425695 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Cancer J Clin. 2017;67(1):7–30. - PubMed
-
- Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology guidelines on renal cell carcinoma: The 2019 update. Eur Urol. 2019;75(5):799–810. - PubMed
-
- Bertero L, Massa F, Metovic J, et al. Eighth Edition of the UICC Classification of Malignant Tumours: An overview of the changes in the pathological TNM classification criteria – what has changed and why? Virchows Archiv. 2018;472(4):519–31. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

