Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains
- PMID: 31194814
- PMCID: PMC6563999
- DOI: 10.1371/journal.pone.0218204
Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains
Abstract
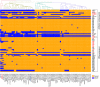
The nosocomial pathogen Acinetobacter baumannii acquired clinical significance due to the rapid development of its multi-drug resistant (MDR) phenotype. A. baumannii strains have the ability to colonize several ecological niches including soil, water, and animals, including humans. They also survive under extremely harsh environmental conditions thriving on rare and recalcitrant carbon compounds. However, the molecular basis behind such extreme adaptability of A. baumannii is unknown. We have therefore determined the complete genome sequence of A. baumannii DS002, which was isolated from agricultural soils, and compared it with 78 complete genome sequences of A. baumannii strains having complete information on the source of their isolation. Interestingly, the genome of A. baumannii DS002 showed high similarity to the genome of A. baumannii SDF isolated from the body louse. The environmental and clinical strains, which do not share a monophyletic origin, showed the existence of a strain-specific unique gene pool that supports niche-specific survival. The strains isolated from infected samples contained a genetic repertoire with a unique gene pool coding for iron acquisition machinery, particularly those required for the biosynthesis of acinetobactin. Interestingly, these strains also contained genes required for biofilm formation. However, such gene sets were either partially or completely missing in the environmental isolates, which instead harbored genes required for alternate carbon catabolism and a TonB-dependent transport system involved in the acquisition of iron via siderophores or xenosiderophores.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Acinetobacter baumannii NCIMB8209: a Rare Environmental Strain Displaying Extensive Insertion Sequence-Mediated Genome Remodeling Resulting in the Loss of Exposed Cell Structures and Defensive Mechanisms.mSphere. 2020 Jul 29;5(4):e00404-20. doi: 10.1128/mSphere.00404-20. mSphere. 2020. PMID: 32727858 Free PMC article.
-
Comparative genomic analysis of Acinetobacter baumannii clinical isolates reveals extensive genomic variation and diverse antibiotic resistance determinants.BMC Genomics. 2014 Dec 22;15(1):1163. doi: 10.1186/1471-2164-15-1163. BMC Genomics. 2014. PMID: 25534766 Free PMC article.
-
Phenotypic and molecular characterization of Acinetobacter baumannii isolates causing lower respiratory infections among ICU patients.Microb Pathog. 2019 Mar;128:75-81. doi: 10.1016/j.micpath.2018.12.023. Epub 2018 Dec 15. Microb Pathog. 2019. PMID: 30562602
-
Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii.Biometals. 2009 Feb;22(1):23-32. doi: 10.1007/s10534-008-9202-3. Epub 2009 Jan 7. Biometals. 2009. PMID: 19130255 Review.
-
Acinetobacter baumannii: evolution of a global pathogen.Pathog Dis. 2014 Aug;71(3):292-301. doi: 10.1111/2049-632X.12125. Epub 2014 Jan 27. Pathog Dis. 2014. PMID: 24376225 Review.
Cited by
-
Phenotypic and Molecular Characterization of Hypervirulent and Multidrug-Resistant Acinetobacter baumannii Isolated from ICU Respiratory Infections.Can J Infect Dis Med Microbiol. 2024 Sep 18;2024:9670708. doi: 10.1155/2024/9670708. eCollection 2024. Can J Infect Dis Med Microbiol. 2024. PMID: 39329052 Free PMC article.
-
Diversity of resistant determinants, virulence factors, and mobile genetic elements in Acinetobacter baumannii from India: A comprehensive in silico genome analysis.Front Cell Infect Microbiol. 2022 Nov 28;12:997897. doi: 10.3389/fcimb.2022.997897. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36519127 Free PMC article.
-
Extensively drug-resistant Acinetobacter baumannii: role of conjugative plasmids in transferring resistance.PeerJ. 2023 Jan 25;11:e14709. doi: 10.7717/peerj.14709. eCollection 2023. PeerJ. 2023. PMID: 36718445 Free PMC article.
-
Outer Membrane Vesicles of Acinetobacter baumannii DS002 Are Selectively Enriched with TonB-Dependent Transporters and Play a Key Role in Iron Acquisition.Microbiol Spectr. 2022 Apr 27;10(2):e0029322. doi: 10.1128/spectrum.00293-22. Epub 2022 Mar 10. Microbiol Spectr. 2022. PMID: 35266817 Free PMC article.
-
Genomic landscape of nosocomial Acinetobacter baumannii: A comprehensive analysis of the resistome, virulome, and mobilome.Sci Rep. 2025 May 25;15(1):18203. doi: 10.1038/s41598-025-03246-7. Sci Rep. 2025. PMID: 40414962 Free PMC article.
References
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2009;48(1):1–12. 10.1086/595011 . - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

