ZEB1 insufficiency causes corneal endothelial cell state transition and altered cellular processing
- PMID: 31194824
- PMCID: PMC6564028
- DOI: 10.1371/journal.pone.0218279
ZEB1 insufficiency causes corneal endothelial cell state transition and altered cellular processing
Abstract
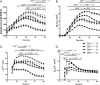
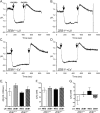
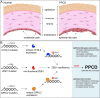
The zinc finger e-box binding homeobox 1 (ZEB1) transcription factor is a master regulator of the epithelial to mesenchymal transition (EMT), and of the reverse mesenchymal to epithelial transition (MET) processes. ZEB1 plays an integral role in mediating cell state transitions during cell lineage specification, wound healing and disease. EMT/MET are characterized by distinct changes in molecular and cellular phenotype that are generally context-independent. Posterior polymorphous corneal dystrophy (PPCD), associated with ZEB1 insufficiency, provides a new biological context in which to understand and evaluate the classic EMT/MET paradigm. PPCD is characterized by a cadherin-switch and transition to an epithelial-like transcriptomic and cellular phenotype, which we study in a cell-based model of PPCD generated using CRISPR-Cas9-mediated ZEB1 knockout in corneal endothelial cells (CEnCs). Transcriptomic and functional studies support the hypothesis that CEnC undergo a MET-like transition in PPCD, termed endothelial to epithelial transition (EnET), and lead to the conclusion that EnET may be considered a corollary to the classic EMT/MET paradigm.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Expression and Function of ZEB1 in the Cornea.Cells. 2021 Apr 16;10(4):925. doi: 10.3390/cells10040925. Cells. 2021. PMID: 33923743 Free PMC article. Review.
-
Transcriptomic Profiling of Posterior Polymorphous Corneal Dystrophy.Invest Ophthalmol Vis Sci. 2017 Jun 1;58(7):3202-3214. doi: 10.1167/iovs.17-21423. Invest Ophthalmol Vis Sci. 2017. PMID: 28654985 Free PMC article.
-
Alterations in GRHL2-OVOL2-ZEB1 axis and aberrant activation of Wnt signaling lead to altered gene transcription in posterior polymorphous corneal dystrophy.Exp Eye Res. 2019 Nov;188:107696. doi: 10.1016/j.exer.2019.107696. Epub 2019 Jun 21. Exp Eye Res. 2019. PMID: 31233731
-
Zeb1 mutant mice as a model of posterior corneal dystrophy.Invest Ophthalmol Vis Sci. 2008 May;49(5):1843-9. doi: 10.1167/iovs.07-0789. Invest Ophthalmol Vis Sci. 2008. PMID: 18436818 Free PMC article.
-
ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance.Cell Cycle. 2015;14(4):481-7. doi: 10.1080/15384101.2015.1006048. Cell Cycle. 2015. PMID: 25607528 Free PMC article. Review.
Cited by
-
Posterior Polymorphous Corneal Dystrophy in a Patient with a Novel ZEB1 Gene Mutation.Int J Mol Sci. 2022 Dec 22;24(1):209. doi: 10.3390/ijms24010209. Int J Mol Sci. 2022. PMID: 36613650 Free PMC article.
-
Diseases of the corneal endothelium.Exp Eye Res. 2021 Apr;205:108495. doi: 10.1016/j.exer.2021.108495. Epub 2021 Feb 14. Exp Eye Res. 2021. PMID: 33596440 Free PMC article. Review.
-
Systematic review of SLC4A11, ZEB1, LOXHD1, and AGBL1 variants in the development of Fuchs' endothelial corneal dystrophy.Front Med (Lausanne). 2023 Jun 27;10:1153122. doi: 10.3389/fmed.2023.1153122. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37441688 Free PMC article.
-
Methamphetamine Induces Systemic Inflammation and Anxiety: The Role of the Gut-Immune-Brain Axis.Int J Mol Sci. 2022 Sep 23;23(19):11224. doi: 10.3390/ijms231911224. Int J Mol Sci. 2022. PMID: 36232524 Free PMC article.
-
Expression and Function of ZEB1 in the Cornea.Cells. 2021 Apr 16;10(4):925. doi: 10.3390/cells10040925. Cells. 2021. PMID: 33923743 Free PMC article. Review.
References
-
- Huang RY, Wong MK, Tan TZ, Kuay KT, Ng AH, Chung VY, et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis. 2013;4:e915 10.1038/cddis.2013.442 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

