Boundaries of eliminated heterochromatin of Tetrahymena are positioned by the DNA-binding protein Ltl1
- PMID: 31194876
- PMCID: PMC6698652
- DOI: 10.1093/nar/gkz504
Boundaries of eliminated heterochromatin of Tetrahymena are positioned by the DNA-binding protein Ltl1
Abstract
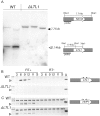
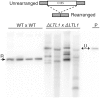
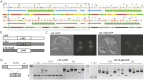
During differentiation of the Tetrahymena thermophila somatic nucleus, its germline-derived DNA undergoes extensive reorganization including the removal of ∼50 Mb from thousands of loci called internal eliminated sequences (IESs). IES-associated chromatin is methylated on lysines 9 and 27 of histone H3, marking newly formed heterochromatin for elimination. To ensure that this reorganized genome maintains essential coding and regulatory sequences, the boundaries of IESs must be accurately defined. In this study, we show that the developmentally expressed protein encoded by Lia3-Like 1 (LTL1) (Ttherm_00499370) is necessary to direct the excision boundaries of particular IESs. In ΔLTL1 cells, boundaries of eliminated loci are aberrant and heterogeneous. The IESs regulated by Ltl1 are distinct from those regulated by the guanine-quadruplex binding Lia3 protein. Ltl1 has a general affinity for double stranded DNA (Kd ∼ 350 nM) and binds specifically to a 50 bp A+T rich sequence flanking each side of the D IES (Kd ∼ 43 nM). Together these data reveal that Ltl1 and Lia3 control different subsets of IESs and that their mechanisms for flanking sequence recognition are distinct.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
A Parallel G Quadruplex-Binding Protein Regulates the Boundaries of DNA Elimination Events of Tetrahymena thermophila.PLoS Genet. 2016 Mar 7;12(3):e1005842. doi: 10.1371/journal.pgen.1005842. eCollection 2016 Mar. PLoS Genet. 2016. PMID: 26950070 Free PMC article.
-
Mutations in Pdd1 reveal distinct requirements for its chromodomain and chromoshadow domain in directing histone methylation and heterochromatin elimination.Eukaryot Cell. 2014 Feb;13(2):190-201. doi: 10.1128/EC.00219-13. Epub 2013 Dec 2. Eukaryot Cell. 2014. PMID: 24297443 Free PMC article.
-
Small-RNA-Mediated Genome-wide trans-Recognition Network in Tetrahymena DNA Elimination.Mol Cell. 2015 Jul 16;59(2):229-42. doi: 10.1016/j.molcel.2015.05.024. Epub 2015 Jun 18. Mol Cell. 2015. PMID: 26095658 Free PMC article.
-
Dynamic nuclear reorganization during genome remodeling of Tetrahymena.Biochim Biophys Acta. 2008 Nov;1783(11):2130-6. doi: 10.1016/j.bbamcr.2008.07.012. Epub 2008 Jul 28. Biochim Biophys Acta. 2008. PMID: 18706458 Free PMC article. Review.
-
DNA elimination in ciliates: transposon domestication and genome surveillance.Annu Rev Genet. 2011;45:227-46. doi: 10.1146/annurev-genet-110410-132432. Epub 2011 Sep 9. Annu Rev Genet. 2011. PMID: 21910632 Review.
Cited by
-
Roles of Noncoding RNAs in Ciliate Genome Architecture.J Mol Biol. 2020 Jul 10;432(15):4186-4198. doi: 10.1016/j.jmb.2019.12.042. Epub 2020 Jan 10. J Mol Biol. 2020. PMID: 31926952 Free PMC article. Review.
-
Programmed genome rearrangements in ciliates.Cell Mol Life Sci. 2020 Nov;77(22):4615-4629. doi: 10.1007/s00018-020-03555-2. Epub 2020 May 27. Cell Mol Life Sci. 2020. PMID: 32462406 Free PMC article. Review.
References
-
- Mochizuki K., Fine N.A., Fujisawa T., Gorovsky M.A.. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002; 110:689–699. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

