Functional characterization of SCN10A variants in several cases of sudden unexplained death
- PMID: 31195250
- PMCID: PMC6625669
- DOI: 10.1016/j.forsciint.2019.05.042
Functional characterization of SCN10A variants in several cases of sudden unexplained death
Abstract
Background: Multiple genome-wide association studies (GWAS) and targeted gene sequencing have identified common variants in SCN10A in cases of PR and QRS duration abnormalities, atrial fibrillation and Brugada syndrome. The New York City Office of Chief Medical Examiner has now also identified five SCN10A variants of uncertain significance in six separate cases within a cohort of 330 sudden unexplained death events. The gene product of SCN10A is the Nav1.8 sodium channel. The purpose of this study was to characterize effects of these variants on Nav1.8 channel function to provide better information for the reclassification of these variants.
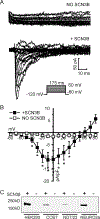
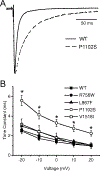
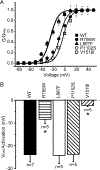
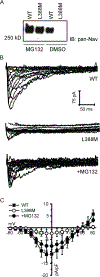
Methods and results: Patch clamp studies were performed to assess effects of the variants on whole-cell Nav1.8 currents. We also performed RNA-seq analysis and immunofluorescence confocal microcopy to determine Nav1.8 expression in heart. We show that four of the five rare 'variants of unknown significance' (L388M, L867F, P1102S and V1518I) are associated with altered functional phenotypes. The R756W variant behaved similar to wild-type under our experimental conditions. We failed to detect Nav1.8 protein expression in immunofluorescence microscopy in rat heart. Furthermore, RNA-seq analysis failed to detect full-length SCN10A mRNA transcripts in human ventricle or mouse specialized cardiac conduction system, suggesting that the effect of Nav1.8 on cardiac function is likely to be extra-cardiac in origin.
Conclusions: We have demonstrated that four of five SCN10A variants of uncertain significance, identified in unexplained death, have deleterious effects on channel function. These data extend the genetic testing of SUD cases, but significantly more clinical evidence is needed to satisfy the criteria needed to associate these variants with the onset of SUD.
Keywords: Channelopathies; Na(+) channels; SCN10A; Sudden unexplained death.
Copyright © 2019 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Variant Intronic Enhancer Controls SCN10A-short Expression and Heart Conduction.Circulation. 2021 Jul 20;144(3):229-242. doi: 10.1161/CIRCULATIONAHA.121.054083. Epub 2021 Apr 29. Circulation. 2021. PMID: 33910361
-
Role of common and rare variants in SCN10A: results from the Brugada syndrome QRS locus gene discovery collaborative study.Cardiovasc Res. 2015 Jun 1;106(3):520-9. doi: 10.1093/cvr/cvv042. Epub 2015 Feb 17. Cardiovasc Res. 2015. PMID: 25691538 Free PMC article.
-
SCN10A/Nav1.8 modulation of peak and late sodium currents in patients with early onset atrial fibrillation.Cardiovasc Res. 2014 Nov 1;104(2):355-63. doi: 10.1093/cvr/cvu170. Epub 2014 Jul 22. Cardiovasc Res. 2014. PMID: 25053638 Free PMC article.
-
Functional significance of channelopathy gene variants in unexplained death.Forensic Sci Med Pathol. 2019 Sep;15(3):437-444. doi: 10.1007/s12024-018-0063-y. Epub 2018 Dec 13. Forensic Sci Med Pathol. 2019. PMID: 30547356 Review.
-
Voltage-gated sodium channels in the mammalian heart.Glob Cardiol Sci Pract. 2014 Dec 31;2014(4):449-63. doi: 10.5339/gcsp.2014.58. eCollection 2014. Glob Cardiol Sci Pract. 2014. PMID: 25780798 Free PMC article. Review.
Cited by
-
Ventricular voltage-gated ion channels: Detection, characteristics, mechanisms, and drug safety evaluation.Clin Transl Med. 2021 Oct;11(10):e530. doi: 10.1002/ctm2.530. Clin Transl Med. 2021. PMID: 34709746 Free PMC article. Review.
-
Role of Genetic Variation in Transcriptional Regulatory Elements in Heart Rhythm.Cells. 2023 Dec 19;13(1):4. doi: 10.3390/cells13010004. Cells. 2023. PMID: 38201209 Free PMC article. Review.
-
Detrimental proarrhythmogenic interaction of Ca2+/calmodulin-dependent protein kinase II and NaV1.8 in heart failure.Nat Commun. 2021 Nov 15;12(1):6586. doi: 10.1038/s41467-021-26690-1. Nat Commun. 2021. PMID: 34782600 Free PMC article.
-
Genetic Factors Underlying Sudden Infant Death Syndrome.Appl Clin Genet. 2021 Feb 15;14:61-76. doi: 10.2147/TACG.S239478. eCollection 2021. Appl Clin Genet. 2021. PMID: 33623412 Free PMC article. Review.
-
Common variants in SCN10A gene associated with Brugada syndrome.Hum Mol Genet. 2021 Dec 27;31(2):157-165. doi: 10.1093/hmg/ddab217. Hum Mol Genet. 2021. PMID: 34312669 Free PMC article.
References
-
- Wang D, Shah KR, Um SY, Eng LS, Zhou B, Lin Y, Mitchell AA, Nicaj L, Prinz M, McDonald TV, Sampson BA and Tang Y. Cardiac channelopathy testing in 274 ethnically diverse sudden unexplained deaths. Forensic Sci Int. 2014;237:90–9. - PubMed
-
- Bagnall RD, Weintraub RG, Ingles J, Duflou J, Yeates L, Lam L, Davis AM, Thompson T, Connell V, Wallace J, Naylor C, Crawford J, Love DR, Hallam L, White J, Lawrence C, Lynch M, Morgan N, James P, du Sart D, Puranik R, Langlois N, Vohra J, Winship I, Atherton J, McGaughran J, Skinner JR and Semsarian C. A Prospective Study of Sudden Cardiac Death among Children and Young Adults. N Engl J Med. 2016;374:2441–52. - PubMed
-
- Brugada J, Brugada R and Brugada P. Channelopathies: a new category of diseases causing sudden death. Herz. 2007;32:185–91. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

