Avian Sarcoma and Leukosis Virus Envelope Glycoproteins Evolve to Broaden Receptor Usage Under Pressure from Entry Competitors †
- PMID: 31195660
- PMCID: PMC6630762
- DOI: 10.3390/v11060519
Avian Sarcoma and Leukosis Virus Envelope Glycoproteins Evolve to Broaden Receptor Usage Under Pressure from Entry Competitors †
Abstract
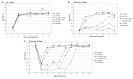
The subgroup A through E avian sarcoma and leukosis viruses (ASLV(A) through ASLV(E)) are a group of highly related alpharetroviruses that have evolved their envelope glycoproteins to use different receptors to enable efficient virus entry due to host resistance and/or to expand host range. Previously, we demonstrated that ASLV(A) in the presence of a competitor to the subgroup A Tva receptor, SUA-rIgG immunoadhesin, evolved to use other receptor options. The selected mutant virus, RCASBP(A)Δ155-160, modestly expanded its use of the Tvb and Tvc receptors and possibly other cell surface proteins while maintaining the binding affinity to Tva. In this study, we further evolved the Δ155-160 virus with the genetic selection pressure of a soluble form of the Tva receptor that should force the loss of Tva binding affinity in the presence of the Δ155-160 mutation. Viable ASLVs were selected that acquired additional mutations in the Δ155-160 Env hypervariable regions that significantly broadened receptor usage to include Tvb and Tvc as well as retaining the use of Tva as a receptor determined by receptor interference assays. A similar deletion in the hr1 hypervariable region of the subgroup C ASLV glycoproteins evolved to broaden receptor usage when selected on Tvc-negative cells.
Keywords: genetic selection inhibiting entry; receptor use expansion; the subgroup A through E avian sarcoma and leukosis viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
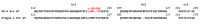


Similar articles
-
Reverse Engineering Provides Insights on the Evolution of Subgroups A to E Avian Sarcoma and Leukosis Virus Receptor Specificity.Viruses. 2019 May 30;11(6):497. doi: 10.3390/v11060497. Viruses. 2019. PMID: 31151254 Free PMC article. Review.
-
Model of the TVA receptor determinants required for efficient infection by subgroup A avian sarcoma and leukosis viruses.J Virol. 2015 Feb;89(4):2136-48. doi: 10.1128/JVI.02339-14. Epub 2014 Dec 3. J Virol. 2015. PMID: 25473063 Free PMC article.
-
Mutations in Both the Surface and Transmembrane Envelope Glycoproteins of the RAV-2 Subgroup B Avian Sarcoma and Leukosis Virus Are Required to Escape the Antiviral Effect of a Secreted Form of the TvbS3 Receptor †.Viruses. 2019 May 31;11(6):500. doi: 10.3390/v11060500. Viruses. 2019. PMID: 31159208 Free PMC article.
-
Two different molecular defects in the Tva receptor gene explain the resistance of two tvar lines of chickens to infection by subgroup A avian sarcoma and leukosis viruses.J Virol. 2004 Dec;78(24):13489-500. doi: 10.1128/JVI.78.24.13489-13500.2004. J Virol. 2004. PMID: 15564460 Free PMC article.
-
Alpharetrovirus envelope-receptor interactions.Curr Top Microbiol Immunol. 2003;281:107-36. doi: 10.1007/978-3-642-19012-4_3. Curr Top Microbiol Immunol. 2003. PMID: 12932076 Review.
Cited by
-
Residues 140-142, 199-200, 222-223, and 262 in the Surface Glycoprotein of Subgroup A Avian Leukosis Virus Are the Key Sites Determining Tva Receptor Binding Affinity and Infectivity.Front Microbiol. 2022 Apr 27;13:868377. doi: 10.3389/fmicb.2022.868377. eCollection 2022. Front Microbiol. 2022. PMID: 35572683 Free PMC article.
-
Reverse Engineering Provides Insights on the Evolution of Subgroups A to E Avian Sarcoma and Leukosis Virus Receptor Specificity.Viruses. 2019 May 30;11(6):497. doi: 10.3390/v11060497. Viruses. 2019. PMID: 31151254 Free PMC article. Review.
-
Single Amino Acids G196 and R198 in hr1 of Subgroup K Avian Leukosis Virus Glycoprotein Are Critical for Tva Receptor Binding.Front Microbiol. 2020 Dec 16;11:596586. doi: 10.3389/fmicb.2020.596586. eCollection 2020. Front Microbiol. 2020. PMID: 33391214 Free PMC article.
-
Rapid adaptive evolution of avian leukosis virus subgroup J in response to biotechnologically induced host resistance.PLoS Pathog. 2024 Aug 15;20(8):e1012468. doi: 10.1371/journal.ppat.1012468. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39146367 Free PMC article.
-
The key amino acid sites 199-205, 269, 319, 321 and 324 of ALV-K env contribute to the weaker replication capacity of ALV-K than ALV-A.Retrovirology. 2022 Aug 24;19(1):19. doi: 10.1186/s12977-022-00598-0. Retrovirology. 2022. PMID: 36002842 Free PMC article.
References
-
- Coffin J.M. Retroviridae: The viruses and their replication. In: Fields B.N., Knipe D.M., editors. Fields Virology. Vol. 2. Lippincott-Raven; Philadelphia, PA, USA: 1996. pp. 1767–1847.
-
- Coffin J.M. Genetic diversity and evolution of retroviruses. Curr. Top. Microbiol. Immunol. 1992;176:143–164. - PubMed
-
- Hunter E. Viral entry and receptors. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1997. pp. 71–120. - PubMed
-
- Weiss R.A. Cellular receptors and viral glycoproteins involved in retrovirus entry. In: Levy J.A., editor. The Retroviruses. Vol. 2. Plenum Press; New York, NY, USA: 1992. pp. 1–108.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

