Organization of DNA in Mammalian Mitochondria
- PMID: 31195723
- PMCID: PMC6600607
- DOI: 10.3390/ijms20112770
Organization of DNA in Mammalian Mitochondria
Abstract
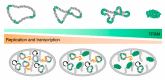
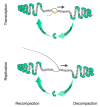
As with all organisms that must organize and condense their DNA to fit within the limited volume of a cell or a nucleus, mammalian mitochondrial DNA (mtDNA) is packaged into nucleoprotein structures called nucleoids. In this study, we first introduce the general modes of DNA compaction, especially the role of the nucleoid-associated proteins (NAPs) that structure the bacterial chromosome. We then present the mitochondrial nucleoid and the main factors responsible for packaging of mtDNA: ARS- (autonomously replicating sequence-) binding factor 2 protein (Abf2p) in yeast and mitochondrial transcription factor A (TFAM) in mammals. We summarize the single-molecule manipulation experiments on mtDNA compaction and visualization of mitochondrial nucleoids that have led to our current knowledge on mtDNA compaction. Lastly, we discuss the possible regulatory role of DNA packaging by TFAM in DNA transactions such as mtDNA replication and transcription.
Keywords: mitochondrial DNA; mtDNA compaction; mtDNA maintenance; mtDNA replication; mtDNA transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

