AutoCryoPicker: an unsupervised learning approach for fully automated single particle picking in Cryo-EM images
- PMID: 31195977
- PMCID: PMC6567647
- DOI: 10.1186/s12859-019-2926-y
AutoCryoPicker: an unsupervised learning approach for fully automated single particle picking in Cryo-EM images
Abstract
Background: An important task of macromolecular structure determination by cryo-electron microscopy (cryo-EM) is the identification of single particles in micrographs (particle picking). Due to the necessity of human involvement in the process, current particle picking techniques are time consuming and often result in many false positives and negatives. Adjusting the parameters to eliminate false positives often excludes true particles in certain orientations. The supervised machine learning (e.g. deep learning) methods for particle picking often need a large training dataset, which requires extensive manual annotation. Other reference-dependent methods rely on low-resolution templates for particle detection, matching and picking, and therefore, are not fully automated. These issues motivate us to develop a fully automated, unbiased framework for particle picking.
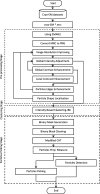
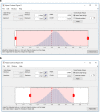
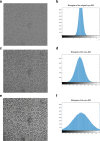
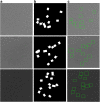
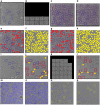
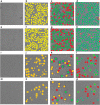
Results: We design a fully automated, unsupervised approach for single particle picking in cryo-EM micrographs. Our approach consists of three stages: image preprocessing, particle clustering, and particle picking. The image preprocessing is based on multiple techniques including: image averaging, normalization, cryo-EM image contrast enhancement correction (CEC), histogram equalization, restoration, adaptive histogram equalization, guided image filtering, and morphological operations. Image preprocessing significantly improves the quality of original cryo-EM images. Our particle clustering method is based on an intensity distribution model which is much faster and more accurate than traditional K-means and Fuzzy C-Means (FCM) algorithms for single particle clustering. Our particle picking method, based on image cleaning and shape detection with a modified Circular Hough Transform algorithm, effectively detects the shape and the center of each particle and creates a bounding box encapsulating the particles.
Conclusions: AutoCryoPicker can automatically and effectively recognize particle-like objects from noisy cryo-EM micrographs without the need of labeled training data or human intervention making it a useful tool for cryo-EM protein structure determination.
Keywords: Clustering; Cryo-EM; Intensity based clustering (IBC); Micrograph; Protein structure determination; Single particle picking.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures












Similar articles
-
A Super-Clustering Approach for Fully Automated Single Particle Picking in Cryo-EM.Genes (Basel). 2019 Aug 30;10(9):666. doi: 10.3390/genes10090666. Genes (Basel). 2019. PMID: 31480377 Free PMC article.
-
DeepCryoPicker: fully automated deep neural network for single protein particle picking in cryo-EM.BMC Bioinformatics. 2020 Nov 9;21(1):509. doi: 10.1186/s12859-020-03809-7. BMC Bioinformatics. 2020. PMID: 33167860 Free PMC article.
-
Automatic post-picking using MAPPOS improves particle image detection from cryo-EM micrographs.J Struct Biol. 2013 May;182(2):59-66. doi: 10.1016/j.jsb.2013.02.008. Epub 2013 Feb 21. J Struct Biol. 2013. PMID: 23454482
-
Artificial intelligence in cryo-EM protein particle picking: recent advances and remaining challenges.Brief Bioinform. 2024 Nov 22;26(1):bbaf011. doi: 10.1093/bib/bbaf011. Brief Bioinform. 2024. PMID: 39820248 Free PMC article. Review.
-
[A review of automatic particle recognition in Cryo-EM images].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010 Oct;27(5):1178-82. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010. PMID: 21089695 Review. Chinese.
Cited by
-
CryoTransformer: A Transformer Model for Picking Protein Particles from Cryo-EM Micrographs.bioRxiv [Preprint]. 2023 Oct 23:2023.10.19.563155. doi: 10.1101/2023.10.19.563155. bioRxiv. 2023. Update in: Bioinformatics. 2024 Mar 4;40(3):btae109. doi: 10.1093/bioinformatics/btae109. PMID: 37961171 Free PMC article. Updated. Preprint.
-
Deep Learning-Based Advances in Protein Structure Prediction.Int J Mol Sci. 2021 May 24;22(11):5553. doi: 10.3390/ijms22115553. Int J Mol Sci. 2021. PMID: 34074028 Free PMC article. Review.
-
Auto3DCryoMap: an automated particle alignment approach for 3D cryo-EM density map reconstruction.BMC Bioinformatics. 2020 Dec 28;21(Suppl 21):534. doi: 10.1186/s12859-020-03885-9. BMC Bioinformatics. 2020. PMID: 33371884 Free PMC article.
-
UPicker: a semi-supervised particle picking transformer method for cryo-EM micrographs.Brief Bioinform. 2024 Nov 22;26(1):bbae636. doi: 10.1093/bib/bbae636. Brief Bioinform. 2024. PMID: 39658205 Free PMC article.
-
A large-scale curated and filterable dataset for cryo-EM foundation model pre-training.Sci Data. 2025 Jun 7;12(1):960. doi: 10.1038/s41597-025-05179-2. Sci Data. 2025. PMID: 40483273 Free PMC article.
References
-
- Doerr, Allison. 2016. “Single-particle cryo-electron microscopy.” Nat Methods 23. https://www.nature.com/articles/nmeth.3700?draft=collection. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

