Entry of Challenge Virus Standard (CVS) -11 into N2a cells via a clathrin-mediated, cholesterol-, dynamin-, pH-dependent endocytic pathway
- PMID: 31196105
- PMCID: PMC6567506
- DOI: 10.1186/s12985-019-1186-9
Entry of Challenge Virus Standard (CVS) -11 into N2a cells via a clathrin-mediated, cholesterol-, dynamin-, pH-dependent endocytic pathway
Abstract
Background: Rabies virus (RABV), a member of Lyssavirus of Rhabdoviridae family, is a kind of negative-strand RNA virus. The zoonosis caused by RABV leads to high mortality in animals and humans. Though with the extensive investigation, the mechanisms of RABV entry into cells have not been well characterized.
Methods: Chemical inhibitors and RNA interference (RNAi) were used to analysis RABV internalization pathway. The expression level of viral N protein was examined by quantitative PCR and western blot, and the virus infection in the cells was visualized by fluorescence microscopy.
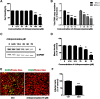
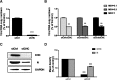
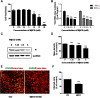
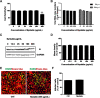
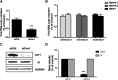
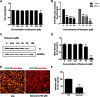
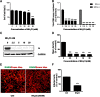
Results: We firstly examined the endocytosis pathway of the challenge virus standard (CVS) -11 strain in N2a cells. Chlorpromazine treatment and knockdown of clathrin heavy chain (CHC) significantly reduced viral entry, which proved clathrin was required. Meanwhile neither nystatin nor knocking down caveolin-1 (Cav1) in N2a cells had an effect on CVS-11 infection, suggesting that caveolae was independent for CVS-11 internalization. And when cholesterol of cell membrane was extracted by MβCD, viral infection was strongly impacted. Additionally by using the specific inhibitor dynasore and ammonium chloride, we verified that dynamin and a low-pH environment were crucial for RABV infection, which was confirmed by confocal microscopy.
Conclusions: Our results demonstrated that CVS-11 entered N2a cells through a clathrin-mediated, cholesterol-, pH-, dynamin-required, and caveolae-independent endocytic pathway.
Keywords: Caveolin-1; Clathrin; Endocytosis; N2a; RABV.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures
Similar articles
-
Hsv-1 Endocytic Entry into a Human Oligodendrocytic Cell Line is Mediated by Clathrin and Dynamin but Not Caveolin.Viruses. 2020 Jul 7;12(7):734. doi: 10.3390/v12070734. Viruses. 2020. PMID: 32645983 Free PMC article.
-
Entry of Classical Swine Fever Virus into PK-15 Cells via a pH-, Dynamin-, and Cholesterol-Dependent, Clathrin-Mediated Endocytic Pathway That Requires Rab5 and Rab7.J Virol. 2016 Sep 29;90(20):9194-208. doi: 10.1128/JVI.00688-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489278 Free PMC article.
-
Bovine Parainfluenza Virus Type 3 (BPIV3) Enters HeLa Cells via Clathrin-Mediated Endocytosis in a Cholesterol- and Dynamin-Dependent Manner.Viruses. 2021 May 31;13(6):1035. doi: 10.3390/v13061035. Viruses. 2021. PMID: 34072688 Free PMC article.
-
Early events in rabies virus infection-Attachment, entry, and intracellular trafficking.Virus Res. 2019 Apr 2;263:217-225. doi: 10.1016/j.virusres.2019.02.006. Epub 2019 Feb 14. Virus Res. 2019. PMID: 30772332 Review.
-
Clathrin-independent endocytosis: an increasing degree of complexity.Histochem Cell Biol. 2018 Aug;150(2):107-118. doi: 10.1007/s00418-018-1678-5. Epub 2018 May 17. Histochem Cell Biol. 2018. PMID: 29774430 Free PMC article. Review.
Cited by
-
Rhabdovirus Infection Is Dependent on Serine/Threonine Kinase AP2-Associated Kinase 1.Life (Basel). 2020 Aug 30;10(9):170. doi: 10.3390/life10090170. Life (Basel). 2020. PMID: 32872567 Free PMC article.
-
Prolonged ethanol exposure modulates constitutive internalization and recycling of 5-HT1A receptors.J Neurochem. 2022 Feb;160(4):469-481. doi: 10.1111/jnc.15564. Epub 2021 Dec 29. J Neurochem. 2022. PMID: 34928513 Free PMC article.
-
Attachment, Entry, and Intracellular Trafficking of Classical Swine Fever Virus.Viruses. 2023 Sep 3;15(9):1870. doi: 10.3390/v15091870. Viruses. 2023. PMID: 37766277 Free PMC article. Review.
-
Schisandra chinensis inhibits the entry of BoHV-1 by blocking PI3K-Akt pathway and enhances the m6A methylation of gD to inhibit the entry of progeny virus.Front Microbiol. 2024 Jul 22;15:1444414. doi: 10.3389/fmicb.2024.1444414. eCollection 2024. Front Microbiol. 2024. PMID: 39104584 Free PMC article.
-
Hsv-1 Endocytic Entry into a Human Oligodendrocytic Cell Line is Mediated by Clathrin and Dynamin but Not Caveolin.Viruses. 2020 Jul 7;12(7):734. doi: 10.3390/v12070734. Viruses. 2020. PMID: 32645983 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

