Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids
- PMID: 31196223
- PMCID: PMC6567389
- DOI: 10.1186/s40478-019-0742-2
Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids
Erratum in
-
Publisher Correction to: Acta Neuropathologica Communications, volume 7.Acta Neuropathol Commun. 2019 Aug 14;7(1):131. doi: 10.1186/s40478-019-0784-5. Acta Neuropathol Commun. 2019. PMID: 31412936 Free PMC article.
Abstract
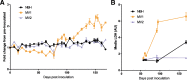
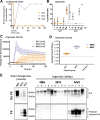
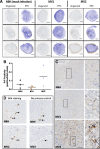
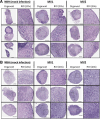
For the transmissible, neurogenerative family of prion diseases, few human models of infection exist and none represent structured neuronal tissue. Human cerebral organoids are self-organizing, three-dimensional brain tissues that can be grown from induced pluripotent stem cells. Organoids can model aspects of neurodegeneration in Alzheimer's Disease and Down's Syndrome, reproducing tau hyperphosphorylation and amyloid plaque pathology. To determine whether organoids could be used to reproduce human prion infection and pathogenesis, we inoculated organoids with two sporadic Creutzfeldt-Jakob Disease prion subtypes. Organoids showed uptake, followed by clearance, of the infectious inoculum. Subsequent re-emergence of prion self-seeding activity indicated de novo propagation. Organoid health assays, prion titer, prion protein electrophoretic mobility and immunohistochemistry demonstrated inoculum-specific differences. Our study shows, for the first time, that cerebral organoids can model aspects of human prion disease and thus offer a powerful system for investigating different human prion subtype pathologies and testing putative therapeutics.
Keywords: CJD; Human cerebral organoid; Induced pluripotent stem cells; Prion; RT-QuIC.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

