Dynamic compartmentalization of purine nucleotide metabolic enzymes at leading edge in highly motile renal cell carcinoma
- PMID: 31196624
- PMCID: PMC6612443
- DOI: 10.1016/j.bbrc.2019.05.190
Dynamic compartmentalization of purine nucleotide metabolic enzymes at leading edge in highly motile renal cell carcinoma
Abstract
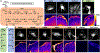
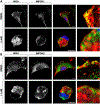
Compartmentalization is vital for biological systems at multiple levels, including biochemical reactions in metabolism. Organelle-based compartments such as mitochondria and peroxisomes sequester the responsible enzymes and increase the efficiency of metabolism while simultaneously protecting the cell from dangerous intermediates, such as radical oxygen species. Recent studies show intracellular nucleotides, such as ATP and GTP, are heterogeneously distributed in cells with high concentrations at the lamellipodial and filopodial projections, or leading edge. However, the intracellular distribution of purine nucleotide enzymes remains unclear. Here, we report the enhanced localization of GTP-biosynthetic enzymes, including inosine monophosphate dehydrogenase (IMPDH isotype 1 and 2), GMP synthase (GMPS), guanylate kinase (GUK1) and nucleoside diphosphate kinase-A (NDPK-A) at the leading edge in renal cell carcinoma cells. They show significant co-localization at the membrane subdomain, and their co-localization pattern at the membrane is distinct from that of the cell body. While other purine nucleotide biosynthetic enzymes also show significant localization at the leading edge, their co-localization pattern with IMPDH is divergent. In contrast, a key glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), predominantly localized in the cytoplasm. Mechanistically, we found that plasma membrane localization of IMPDH isozymes requires active actin polymerization. Our results demonstrate the formation of a discrete metabolic compartment for localized purine biosynthesis at the leading edge, which may promote localized nucleotide metabolism for cell migration and metastasis in cancers.
Keywords: GMPS; GUK1; IMPDH; Metabolic compartmentalization; Salvage purine synthesis; de novo purine synthesis.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures
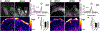

Similar articles
-
Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis.J Biol Chem. 2015 Mar 13;290(11):6705-13. doi: 10.1074/jbc.M114.628701. Epub 2015 Jan 20. J Biol Chem. 2015. PMID: 25605736 Free PMC article.
-
The CBS subdomain of inosine 5'-monophosphate dehydrogenase regulates purine nucleotide turnover.Mol Microbiol. 2008 Apr;68(2):342-59. doi: 10.1111/j.1365-2958.2008.06153.x. Epub 2008 Feb 26. Mol Microbiol. 2008. PMID: 18312263 Free PMC article.
-
Regulation of GTP biosynthesis.Adv Enzyme Regul. 1992;32:57-69. doi: 10.1016/0065-2571(92)90008-n. Adv Enzyme Regul. 1992. PMID: 1353938
-
Enzymes involved in purine metabolism--a review of histochemical localization and functional implications.Histol Histopathol. 1999 Oct;14(4):1321-40. doi: 10.14670/HH-14.1321. Histol Histopathol. 1999. PMID: 10506947 Review.
-
Mechanism of action of mycophenolate mofetil.Ther Drug Monit. 1995 Dec;17(6):681-4. doi: 10.1097/00007691-199512000-00023. Ther Drug Monit. 1995. PMID: 8588241 Review.
Cited by
-
Structure, Folding and Stability of Nucleoside Diphosphate Kinases.Int J Mol Sci. 2020 Sep 16;21(18):6779. doi: 10.3390/ijms21186779. Int J Mol Sci. 2020. PMID: 32947863 Free PMC article. Review.
-
Depletion of nuclear cytoophidia in Alzheimer's disease.Free Neuropathol. 2025 Mar 7;6:8. doi: 10.17879/freeneuropathology-2025-6282. eCollection 2025 Jan. Free Neuropathol. 2025. PMID: 40070795 Free PMC article.
-
IMPDH2 and HPRT expression and a prognostic significance in preoperative and postoperative patients with osteosarcoma.Sci Rep. 2021 May 25;11(1):10887. doi: 10.1038/s41598-021-90456-4. Sci Rep. 2021. PMID: 34035425 Free PMC article.
-
Redox dynamics in seeds of Acer spp: unraveling adaptation strategies of different seed categories.Front Plant Sci. 2024 Jul 24;15:1430695. doi: 10.3389/fpls.2024.1430695. eCollection 2024. Front Plant Sci. 2024. PMID: 39114470 Free PMC article.
-
The bioenergetics of neuronal morphogenesis and regeneration: Frontiers beyond the mitochondrion.Dev Neurobiol. 2020 Jul;80(7-8):263-276. doi: 10.1002/dneu.22776. Epub 2020 Sep 27. Dev Neurobiol. 2020. PMID: 32750228 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

