Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells
- PMID: 31196865
- PMCID: PMC6672052
- DOI: 10.1101/gad.327494.119
Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells
Abstract
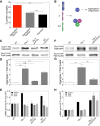
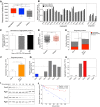
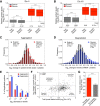
Aneuploidy, a condition characterized by chromosome gains and losses, causes reduced fitness and numerous cellular stresses, including increased protein aggregation. Here, we identify protein complex stoichiometry imbalances as a major cause of protein aggregation in aneuploid cells. Subunits of protein complexes encoded on excess chromosomes aggregate in aneuploid cells, which is suppressed when expression of other subunits is coordinately altered. We further show that excess subunits are either degraded or aggregate and that protein aggregation is nearly as effective as protein degradation at lowering levels of excess proteins. Our study explains why proteotoxic stress is a universal feature of the aneuploid state and reveals protein aggregation as a form of dosage compensation to cope with disproportionate expression of protein complex subunits.
Keywords: aneuploidy; protein aggregation; protein homeostasis.
© 2019 Brennan et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Comment in
-
Dosage compensation plans: protein aggregation provides additional insurance against aneuploidy.Genes Dev. 2019 Aug 1;33(15-16):1027-1030. doi: 10.1101/gad.329383.119. Genes Dev. 2019. PMID: 31371460 Free PMC article.
