A Mitochondrial Transcription Termination Factor, ZmSmk3, Is Required for nad1 Intron4 and nad4 Intron1 Splicing and Kernel Development in Maize
- PMID: 31196888
- PMCID: PMC6686911
- DOI: 10.1534/g3.119.400265
A Mitochondrial Transcription Termination Factor, ZmSmk3, Is Required for nad1 Intron4 and nad4 Intron1 Splicing and Kernel Development in Maize
Abstract
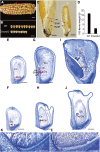
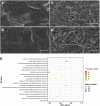
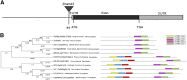
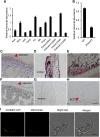
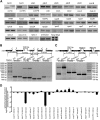
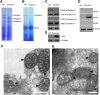
The expression systems of the mitochondrial genes are derived from their bacterial ancestors, but have evolved many new features in their eukaryotic hosts. Mitochondrial RNA splicing is a complex process regulated by families of nucleus-encoded RNA-binding proteins, few of which have been characterized in maize (Zea mays L.). Here, we identified the Zea mays small kernel 3 (Zmsmk3) candidate gene, which encodes a mitochondrial transcription termination factor (mTERF) containing two mTERF motifs, which is conserved in monocotyledon; and the target introns were also quite conserved during evolution between monocotyledons and dicotyledons. The mutations of Zmsmk3 led to arrested embryo and endosperm development, resulting in small kernels. A transcriptome of 12 days after pollination endosperm analysis revealed that the starch biosynthetic pathway and the zein gene family were down-regulated in the Zmsmk3 mutant kernels. ZmSMK3 is localized in mitochondria. The reduced expression of ZmSmk3 in the mutant resulted in the splicing deficiency of mitochondrial nad4 intron1 and nad1 intron4, causing a reduction in complex I assembly and activity, impairing mitochondria structure and activating the alternative respiratory pathway. So, the results suggest that ZmSMK3 is required for the splicing of nad4 intron 1 and nad1 intron 4, complex I assembly and kernel development in maize.
Keywords: Zea mays; embryo; endosperm; intron splicing; mitochondrial.
Copyright © 2019 Pan et al.
Figures
Similar articles
-
Research Progress in the Molecular Functions of Plant mTERF Proteins.Cells. 2021 Jan 21;10(2):205. doi: 10.3390/cells10020205. Cells. 2021. PMID: 33494215 Free PMC article. Review.
-
The Mitochondrial Pentatricopeptide Repeat Protein PPR18 Is Required for the cis-Splicing of nad4 Intron 1 and Essential to Seed Development in Maize.Int J Mol Sci. 2020 Jun 5;21(11):4047. doi: 10.3390/ijms21114047. Int J Mol Sci. 2020. PMID: 32516991 Free PMC article.
-
The novel E-subgroup pentatricopeptide repeat protein DEK55 is responsible for RNA editing at multiple sites and for the splicing of nad1 and nad4 in maize.BMC Plant Biol. 2020 Dec 9;20(1):553. doi: 10.1186/s12870-020-02765-x. BMC Plant Biol. 2020. PMID: 33297963 Free PMC article.
-
Dek35 Encodes a PPR Protein that Affects cis-Splicing of Mitochondrial nad4 Intron 1 and Seed Development in Maize.Mol Plant. 2017 Mar 6;10(3):427-441. doi: 10.1016/j.molp.2016.08.008. Epub 2016 Sep 3. Mol Plant. 2017. PMID: 27596292
-
EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize.J Exp Bot. 2017 Jul 20;68(16):4571-4581. doi: 10.1093/jxb/erx212. J Exp Bot. 2017. PMID: 28981788 Free PMC article.
Cited by
-
Genomic Scan of Male Fertility Restoration Genes in a 'Gülzow' Type Hybrid Breeding System of Rye (Secale cereale L.).Int J Mol Sci. 2021 Aug 27;22(17):9277. doi: 10.3390/ijms22179277. Int J Mol Sci. 2021. PMID: 34502186 Free PMC article.
-
Importance of pre-mRNA splicing and its study tools in plants.Adv Biotechnol (Singap). 2024 Feb 8;2(1):4. doi: 10.1007/s44307-024-00009-9. Adv Biotechnol (Singap). 2024. PMID: 39883322 Free PMC article. Review.
-
Research Progress of Group II Intron Splicing Factors in Land Plant Mitochondria.Genes (Basel). 2024 Jan 28;15(2):176. doi: 10.3390/genes15020176. Genes (Basel). 2024. PMID: 38397166 Free PMC article. Review.
-
Residual effect of defeated stripe rust resistance genes/QTLs in bread wheat against prevalent pathotypes of Puccinia striiformis f. sp. tritici.PLoS One. 2022 Apr 1;17(4):e0266482. doi: 10.1371/journal.pone.0266482. eCollection 2022. PLoS One. 2022. PMID: 35363829 Free PMC article.
-
Research Progress in the Molecular Functions of Plant mTERF Proteins.Cells. 2021 Jan 21;10(2):205. doi: 10.3390/cells10020205. Cells. 2021. PMID: 33494215 Free PMC article. Review.
References
-
- Babiychuk E., Vandepoele K., Wissing J., Garcia-Diaz M., De Rycke R. et al. , 2011. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. USA 108: 6674–6679. 10.1073/pnas.1103442108 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
