Targeting IRAK4 disrupts inflammatory pathways and delays tumor development in chronic lymphocytic leukemia
- PMID: 31197259
- PMCID: PMC8075947
- DOI: 10.1038/s41375-019-0507-8
Targeting IRAK4 disrupts inflammatory pathways and delays tumor development in chronic lymphocytic leukemia
Abstract
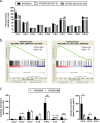
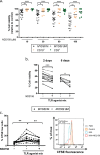
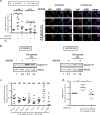
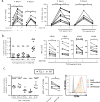
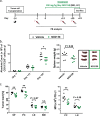
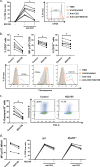
Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a critical role in Toll-like receptor (TLR) signal transduction and innate immune responses. Recruitment and subsequent activation of IRAK4 upon TLR stimulation is mediated by the myeloid differentiation primary response 88 (MYD88) adaptor protein. Around 3% of chronic lymphocytic leukemia (CLL) patients have activating mutations of MYD88, a driver mutation in this disease. Here, we studied the effects of TLR activation and the pharmacological inhibition of IRAK4 with ND2158, an IRAK4 competitive inhibitor, as a therapeutic approach in CLL. Our in vitro studies demonstrated that ND2158 preferentially killed CLL cells in a dose-dependent manner. We further observed a decrease in NF-κB and STAT3 signaling, cytokine secretion, proliferation and migration of primary CLL cells from MYD88-mutated and -unmutated cases. In the Eµ-TCL1 adoptive transfer mouse model of CLL, ND2158 delayed tumor progression and modulated the activity of myeloid and T cells. Our findings show the importance of TLR signaling in CLL development and suggest IRAK4 as a therapeutic target for this disease.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Interleukin-1 receptor-associated kinase 4 inhibitor interrupts toll-like receptor signalling and sensitizes chronic lymphocytic leukaemia cells to apoptosis.Br J Haematol. 2020 May;189(3):475-488. doi: 10.1111/bjh.16386. Epub 2020 Feb 14. Br J Haematol. 2020. PMID: 32057093
-
Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a dual role in myddosome formation and Toll-like receptor signaling.J Biol Chem. 2018 Sep 28;293(39):15195-15207. doi: 10.1074/jbc.RA118.003314. Epub 2018 Aug 3. J Biol Chem. 2018. PMID: 30076215 Free PMC article.
-
MYD88 L265P mutations identify a prognostic gene expression signature and a pathway for targeted inhibition in CLL.Br J Haematol. 2019 Mar;184(6):925-936. doi: 10.1111/bjh.15714. Epub 2018 Dec 9. Br J Haematol. 2019. PMID: 30537114
-
NF-κB activation in chronic lymphocytic leukemia: A point of convergence of external triggers and intrinsic lesions.Semin Cancer Biol. 2016 Aug;39:40-8. doi: 10.1016/j.semcancer.2016.07.005. Epub 2016 Aug 1. Semin Cancer Biol. 2016. PMID: 27491692 Review.
-
Targeting inflammatory pathways in chronic lymphocytic leukemia.Crit Rev Oncol Hematol. 2013 Dec;88(3):655-66. doi: 10.1016/j.critrevonc.2013.07.011. Epub 2013 Aug 12. Crit Rev Oncol Hematol. 2013. PMID: 23941728 Free PMC article. Review.
Cited by
-
Perspectives on Precision Medicine in Chronic Lymphocytic Leukemia: Targeting Recurrent Mutations-NOTCH1, SF3B1, MYD88, BIRC3.J Clin Med. 2021 Aug 22;10(16):3735. doi: 10.3390/jcm10163735. J Clin Med. 2021. PMID: 34442029 Free PMC article. Review.
-
Deep Learning-Driven Library Design for the De Novo Discovery of Bioactive Thiopeptides.ACS Cent Sci. 2023 Nov 7;9(11):2150-2160. doi: 10.1021/acscentsci.3c00957. eCollection 2023 Nov 22. ACS Cent Sci. 2023. PMID: 38033794 Free PMC article.
-
IRAK4 is an immunological checkpoint in neuropsychiatric systemic lupus erythematosus.Sci Rep. 2024 Jul 16;14(1):16393. doi: 10.1038/s41598-024-63567-x. Sci Rep. 2024. PMID: 39014006 Free PMC article.
-
The Role of Bruton's Kinase Inhibitors in Chronic Lymphocytic Leukemia: Current Status and Future Directions.Cancers (Basel). 2022 Feb 2;14(3):771. doi: 10.3390/cancers14030771. Cancers (Basel). 2022. PMID: 35159041 Free PMC article. Review.
-
Application and prospect of targeting innate immune sensors in the treatment of autoimmune diseases.Cell Biosci. 2022 May 26;12(1):68. doi: 10.1186/s13578-022-00810-w. Cell Biosci. 2022. PMID: 35619184 Free PMC article. Review.
References
-
- Ruprecht CR, Lanzavecchia A. Toll-like receptor stimulation as a third signal required for activation of human naive B cells. Eur J Immunol. 2006;36:810–6. - PubMed
-
- Ntoufa S, Vilia MG, Stamatopoulos K, Ghia P, Muzio M. Toll-like receptors signaling: a complex network for NF-κB activation in B-cell lymphoid malignancies. Semin Cancer Biol. 2016;39:15–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

