iScore: a novel graph kernel-based function for scoring protein-protein docking models
- PMID: 31199455
- PMCID: PMC6956772
- DOI: 10.1093/bioinformatics/btz496
iScore: a novel graph kernel-based function for scoring protein-protein docking models
Abstract
Motivation: Protein complexes play critical roles in many aspects of biological functions. Three-dimensional (3D) structures of protein complexes are critical for gaining insights into structural bases of interactions and their roles in the biomolecular pathways that orchestrate key cellular processes. Because of the expense and effort associated with experimental determinations of 3D protein complex structures, computational docking has evolved as a valuable tool to predict 3D structures of biomolecular complexes. Despite recent progress, reliably distinguishing near-native docking conformations from a large number of candidate conformations, the so-called scoring problem, remains a major challenge.
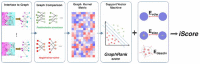
Results: Here we present iScore, a novel approach to scoring docked conformations that combines HADDOCK energy terms with a score obtained using a graph representation of the protein-protein interfaces and a measure of evolutionary conservation. It achieves a scoring performance competitive with, or superior to, that of state-of-the-art scoring functions on two independent datasets: (i) Docking software-specific models and (ii) the CAPRI score set generated by a wide variety of docking approaches (i.e. docking software-non-specific). iScore ranks among the top scoring approaches on the CAPRI score set (13 targets) when compared with the 37 scoring groups in CAPRI. The results demonstrate the utility of combining evolutionary, topological and energetic information for scoring docked conformations. This work represents the first successful demonstration of graph kernels to protein interfaces for effective discrimination of near-native and non-native conformations of protein complexes.
Availability and implementation: The iScore code is freely available from Github: https://github.com/DeepRank/iScore (DOI: 10.5281/zenodo.2630567). And the docking models used are available from SBGrid: https://data.sbgrid.org/dataset/684).
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures


References
-
- Aloy P., Russell R.B. (2006) Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol., 7, 188–197. - PubMed
-
- Andreani J., Guerois R. (2014) Evolution of protein interactions: from interactomes to interfaces. Arch. Biochem. Biophys., 554, 65–75. - PubMed
-
- Andreani J. et al. (2013) InterEvScore: a novel coarse-grained interface scoring function using a multi-body statistical potential coupled to evolution. Bioinformatics, 29, 1742–1749. - PubMed
-
- Borgwardt K.M. et al. (2005) Protein function prediction via graph kernels. Bioinformatics, 21, i47–i56. - PubMed

